Play all audios:
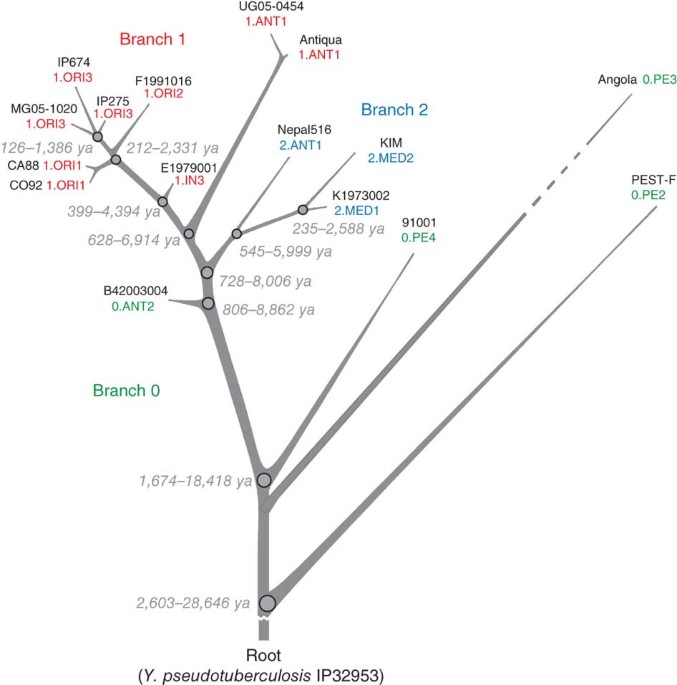
ABSTRACT Plague is a pandemic human invasive disease caused by the bacterial agent _Yersinia pestis_. We here report a comparison of 17 whole genomes of _Y. pestis_ isolates from global
sources. We also screened a global collection of 286 _Y. pestis_ isolates for 933 SNPs using Sequenom MassArray SNP typing. We conducted phylogenetic analyses on this sequence variation
dataset, assigned isolates to populations based on maximum parsimony and, from these results, made inferences regarding historical transmission routes. Our phylogenetic analysis suggests
that _Y. pestis_ evolved in or near China and spread through multiple radiations to Europe, South America, Africa and Southeast Asia, leading to country-specific lineages that can be traced
by lineage-specific SNPs. All 626 current isolates from the United States reflect one radiation, and 82 isolates from Madagascar represent a second radiation. Subsequent local microevolution
of _Y. pestis_ is marked by sequential, geographically specific SNPs. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution
ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article *
Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn
about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS PLAGUED BY A CRYPTIC CLOCK: INSIGHT AND ISSUES FROM THE GLOBAL PHYLOGENY
OF _YERSINIA PESTIS_ Article Open access 19 January 2023 _YERSINIA PESTIS_ GENOMES REVEAL PLAGUE IN BRITAIN 4000 YEARS AGO Article Open access 30 May 2023 GENOMIC DIVERSITY OF _YERSINIA
PESTIS_ FROM YUNNAN PROVINCE, CHINA, IMPLIES A POTENTIAL COMMON ANCESTOR AS THE SOURCE OF TWO PLAGUE EPIDEMICS Article Open access 15 August 2023 ACCESSION CODES ACCESSIONS ARRAYEXPRESS *
E-MTAB-213 REFERENCES * Linz, B. et al. An African origin for the intimate association between humans and _Helicobacter pylori_. _Nature_ 445, 915–918 (2007). Article Google Scholar *
Diamond, J. _Guns, Germs and Steel_ 1–480 (Jonathan Cape, London, UK, 1997). * Stenseth, N.C. et al. Plague: past, present, and future. _PLoS Med._ 5, e3 (2008). Article Google Scholar *
Keeling, M.J. & Gilligan, C.A. Metapopulation dynamics of bubonic plague. _Nature_ 407, 903–906 (2000). Article CAS Google Scholar * Pollitzer, R. Plague studies. 1. A summary of the
history and survey of the present distribution of the disease. _Bull. World Health Organ._ 4, 475–533 (1951). CAS PubMed PubMed Central Google Scholar * Devignat, R. Variétés de l′espèce
_Pasteurella pestis_. Nouvelle hypothèse. _Bull. World Health Organ._ 4, 247–263 (1951). CAS PubMed PubMed Central Google Scholar * Wu, L.T. Chapter I: historical aspects. in _Plague: A
Manual for Medical and Public Health Workers_ 1–55 (Weishengshu, National Quarantine Service, Shanghai, China, 1936). * Anisimov, A.P., Lindler, L.E. & Pier, G.B. Intraspecific
diversity of _Yersinia pestis_. _Clin. Microbiol. Rev._ 17, 434–464 (2004). Article CAS Google Scholar * Achtman, M. Evolution, population structure and phylogeography of genetically
monomorphic bacterial pathogens. _Annu. Rev. Microbiol._ 62, 53–70 (2008). Article CAS Google Scholar * Achtman, M. et al. Microevolution and history of the plague bacillus, _Yersinia
pestis_. _Proc. Natl. Acad. Sci. USA_ 101, 17837–17842 (2004). Article CAS Google Scholar * Brygoo, E.R. Epidemiologie de la peste à Madagascar. _Archive de l′lnstitut Pasteur de
Madagascar_ 35, 7–147 (1966). Google Scholar * Girard, G. Immunity in plague. Acquisitions supplied by 30 years of work on the “_Pasteurella_ pestis EV” (Girard and Robic) strain. _Biol.
Med. (Paris)_ 52, 631–731 (1963). CAS Google Scholar * Li, Y. et al. Genotyping and phylogenetic analysis of _Yersinia pestis_ by MLVA: insights into the worldwide expansion of Central
Asia plague foci. _PLoS ONE_ 4, e6000 (2009). Article Google Scholar * Haensch, S. et al. Distinct clones of _Yersinia pestis_ caused the Black Death. _PLoS Pathog_ 6, e1001134 (2010).
Article Google Scholar * Silkroad Foundation. The Bridge between Eastern and Western Cultures. (2009)〈http://www.silkroadfoundation.org/toc/index.html〉. * Levathes, L. _When China Ruled
the Seas: The Treasure Fleet of the Dragon Throne, 1405–1433_ 1–252 (Oxford University Press, New York, 1996). * Link, V.B. A history of plague in United States of America. _Public Health
Monogr._ 26, 1–120 (1955). CAS PubMed Google Scholar * Yu, H.L. & Christakos, G. Spatiotemporal modelling and mapping of the bubonic plague epidemic in India. _Int. J. Health Geogr._
5, 12 (2006). Article Google Scholar * Del Rio, A., Zegers, R., Boza, R.D. & Montero, L. Informe sobre la epidemia de peste bubonica. _La Chilena di Hijiene_ IX, 1–7 (1904). Google
Scholar * Nocht, B. & Giemsa G. Ueber die Vernichtung von Ratten an Bord von Schiffen als Massregel gegen die Einschleppung der Pest. _Arbeiten aus dem Kaiserlichen Gesundheitsamte_ XX,
6 (1903). * Lauzeral, P. & Millischer, P. A propos de la filiation de deux cas. _Bull. Soc. Pathol. Exot._ 25, 935–941 (1932). Google Scholar * Drummond, A.J. & Rambaut, A. BEAST:
Bayesian evolutionary analysis by sampling trees. _BMC Evol. Biol._ 7, 214 (2007). Article Google Scholar * Adjemian, J.Z., Foley, P., Gage, K.L. & Foley, J.E. Initiation and spread of
traveling waves of plague, _Yersinia pestis_, in the western United States. _Am. J. Trop. Med. Hyg._ 76, 365–375 (2007). Article Google Scholar * Guiyoule, A. et al. Recent emergence of
new variants of _Yersinia pestis_ in Madagascar. _J. Clin. Microbiol._ 35, 2826–2833 (1997). CAS PubMed PubMed Central Google Scholar * Zhou, D. et al. DNA microarray analysis of genome
dynamics in _Yersinia pestis_: insights into bacterial genome microevolution and niche adaptation. _J. Bacteriol._ 186, 5138–5146 (2004). Article CAS Google Scholar * Eppinger, M. et al.
Draft genome sequences of _Yersinia pestis_ isolates from natural foci of endemic plague in China. _J. Bacteriol._ 191, 7628–7629 (2009). Article CAS Google Scholar * Eppinger, M. et al.
Genome sequence of the deep-rooted _Yersinia pestis_ strain Angola reveals new insights into the evolution and pangenome of the plague bacterium. _J. Bacteriol._ 192, 1685–1699 (2010).
Article CAS Google Scholar * Auerbach, R.K. et al. _Yersinia pestis_ evolution on a small timescale: comparison of whole genome sequences from North America. _PLoS ONE_ 2, e770 (2007).
Article Google Scholar * Touchman, J.W. et al. A North American _Yersinia pestis_ draft genome sequence: SNPs and phylogenetic analysis. _PLoS ONE_ 2, e220 (2007). Article Google Scholar
* Garcia, E. et al. Pestoides F, an atypical _Yersinia pestis_ strain from the former Soviet Union. _Adv. Exp. Med. Biol._ 603, 17–22 (2007). Article Google Scholar * Song, Y. et al.
Complete genome sequence of _Yersinia pestis_ strain 91001, an isolate avirulent to humans. _DNA Res._ 11, 179–197 (2004). Article CAS Google Scholar * Chain, P.S. et al. Complete genome
sequence of _Yersinia pestis_ strains Antiqua and Nepal516: evidence of gene reduction in an emerging pathogen. _J. Bacteriol._ 188, 4453–4463 (2006). Article Google Scholar * Parkhill, J.
et al. Genome sequence of _Yersinia pestis_, the causative agent of plague. _Nature_ 413, 523–527 (2001). Article CAS Google Scholar * Deng, W. et al. Genome sequence of _Yersinia
pestis_ KIM. _J. Bacteriol._ 184, 4601–4611 (2002). Article CAS Google Scholar * Chain, P.S.G. et al. Insights into the genome evolution of _Yersinia pestis_ through whole genome
comparison with _Yersinia pseudotuberculosis_. _Proc. Natl. Acad. Sci. USA_ 101, 13826–13831 (2004). Article CAS Google Scholar * Welch, T.J. et al. Multiple antimicrobial resistance in
plague: an emerging public health risk. _PLoS ONE_ 2, e309 (2007). Article Google Scholar * Galimand, M. et al. Multidrug resistance in _Yersinia pestis_ mediated by a transferable
plasmid. _N. Engl. J. Med._ 337, 677–680 (1997). Article CAS Google Scholar * Pearson, T., Okinaka, R.T., Foster, J.T. & Keim, P. Phylogenetic understanding of clonal populations in
an era of whole genome sequencing. _Infect. Genet. Evol._ 9, 1010–1019 (2009). Article CAS Google Scholar * Roumagnac, P. et al. Evolutionary history of _Salmonella_ Typhi. _Science_ 314,
1301–1304 (2006). Article CAS Google Scholar * Nelson, K.E. et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of _Thermotoga maritima_. _Nature_
399, 323–329 (1999). Article CAS Google Scholar * Eppinger, M. et al. The complete genome sequence of _Yersinia pseudotuberculosis_ IP31758, the causative Agent of Far East scarlet-like
fever. _PLoS Genet._ 3, e142 (2007). Article Google Scholar * Huson, D.H. et al. Design of a compartmentalized shotgun assembler for the human genome. _Bioinformatics_ 17 (Suppl 1),
S132–S139 (2001). Article Google Scholar * Holt, K.E. et al. High-throughput sequencing provides insights into genome variation and evolution in _Salmonella_ Typhi. _Nat. Genet._ 40,
987–993 (2008). Article CAS Google Scholar * Lowder, B.V. et al. Recent human-to-poultry host jump, adaptation, and pandemic spread of _Staphylococcus aureus_. _Proc. Natl. Acad. Sci.
USA_ 106, 19545–19550 (2009). Article CAS Google Scholar * Kurtz, S. et al. Versatile and open software for comparing large genomes. _Genome Biol._ 5, R12 (2004). Article Google Scholar
* Vogler, A.J. et al. Assays for the rapid and specific identification of North American _Yersinia pestis_ and the common laboratory strain CO92. _Biotechniques_ 44, 201 203–204, 207
(2008). Article CAS Google Scholar * Vogler, A.J. et al. Phylogeography of _Francisella tularensis_: global expansion of a highly fit clone. _J. Bacteriol._ 191, 2474–2484 (2009). Article
CAS Google Scholar Download references ACKNOWLEDGEMENTS We gratefully acknowledge technical assistance by R. Nera and A. Doyle and helpful comments from A. Rambaut and D. Falush. Support
was provided by grants from the German Army Medical Corps (MSAB15A013) and the Science Foundation of Ireland (05/FE1/B882) to M.A., the National Key Program for Infectious Diseases of China
(2008ZX10004-009) and the State Key Development Program for Basic Research of China (2009CB522600) to R.Y. and the US Department of Homeland Security (NBCH2070001; HSHQDC-08-C00158) and US
National Institutes of Health (AI065359) to P.K. and D.M.W. Whole genome sequencing of _Y. pestis_ strains IP275, MG05-1020 and UG05-045 was supported by federal funds from the National
Institute of Allergy and Infectious Diseases, US National Institutes of Health, Department of Health and Human Services (N01 AI-30071), and sequencing of IP674 was supported by funding for
Sanger Institute Pathogen Genomics by the Wellcome Trust. Genomic DNA of _Y. pestis_ MG05-1020 was kindly provided by S. Bearden and M. Schriefer (Centers for Disease Control and Prevention,
Fort Collins, Colorado, USA). AUTHOR INFORMATION Author notes * Giovanna Morelli, Yajun Song, Camila J Mazzoni & Mark Eppinger Present address: These authors contributed equally to this
work. Present addresses: Max-Planck-Institut für molekulare Genetik, Berlin, Germany (G.M. and B.K.), Berlin Center for Genomics in Biodiversity Research, Berlin, Germany (C.J.M.) and
Max-Delbrück-Centrum für molekulare Medizin (MDC) Berlin-Buch, Berlin, Germany (M.F.)., AUTHORS AND AFFILIATIONS * Department of Molecular Biology, Max-Planck-Institut für
Infektionsbiologie, Berlin, Germany Giovanna Morelli, Camila J Mazzoni, Philippe Roumagnac, Mirjam Feldkamp, Barica Kusecek, Thierry Wirth & Mark Achtman * State Key Laboratory of
Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing, China Yajun Song, Yanjun Li, Yujun Cui & Ruifu Yang * Environmental Research Institute, University
College Cork, Cork, Ireland Yajun Song, Camila J Mazzoni & Mark Achtman * Institute for Genome Sciences, University of Maryland School of Medicine, Baltimore, Maryland, USA Mark Eppinger
& Jacques Ravel * Centre de Coopération Internationale en Recherche Agronomique pour le Développement, Mixte Research Unit Biology and Genetics of Plant/Pathogen Interactions (UMR
BGPI), Montpellier, France Philippe Roumagnac * Department of Biological Sciences, Northern Arizona University, Flagstaff, Arizona, USA David M Wagner, Amy J Vogler & Paul Keim * The
Wellcome Trust Sanger Institute, The Wellcome Trust Genome Campus, Hinxton, Cambridge, UK Nicholas R Thomson * Medical Research Council (MRC) Centre for Outbreak Analysis and Modeling,
Imperial College Faculty of Medicine, London, UK Thibaut Jombart & Francois Balloux * Department of Systematics and Evolution UMR-CNRS 7205, Muséum National d′Histoire Naturelle–Ecole
Pratique des Hautes Etudes, Paris, France Raphael Leblois & Thierry Wirth * Institute of Human Genetics, German Research Center for Environmental Health, Neuherberg, Germany Peter
Lichtner * Unité Peste, Institut Pasteur de Madagascar, Madagascar Lila Rahalison * Division of Vector-Borne Infectious Diseases, Centers for Disease Control and Prevention, Fort Collins,
Colorado, USA Jeannine M Petersen * Pathogen Genomics Division, Translational Genomics Research Institute, Phoenix, Arizona, USA Paul Keim * Institut Pasteur, Yersinia Research Unit, Paris,
France Elisabeth Carniel * Department of Microbiology, University College Cork, Cork, Ireland Mark Achtman Authors * Giovanna Morelli View author publications You can also search for this
author inPubMed Google Scholar * Yajun Song View author publications You can also search for this author inPubMed Google Scholar * Camila J Mazzoni View author publications You can also
search for this author inPubMed Google Scholar * Mark Eppinger View author publications You can also search for this author inPubMed Google Scholar * Philippe Roumagnac View author
publications You can also search for this author inPubMed Google Scholar * David M Wagner View author publications You can also search for this author inPubMed Google Scholar * Mirjam
Feldkamp View author publications You can also search for this author inPubMed Google Scholar * Barica Kusecek View author publications You can also search for this author inPubMed Google
Scholar * Amy J Vogler View author publications You can also search for this author inPubMed Google Scholar * Yanjun Li View author publications You can also search for this author inPubMed
Google Scholar * Yujun Cui View author publications You can also search for this author inPubMed Google Scholar * Nicholas R Thomson View author publications You can also search for this
author inPubMed Google Scholar * Thibaut Jombart View author publications You can also search for this author inPubMed Google Scholar * Raphael Leblois View author publications You can also
search for this author inPubMed Google Scholar * Peter Lichtner View author publications You can also search for this author inPubMed Google Scholar * Lila Rahalison View author publications
You can also search for this author inPubMed Google Scholar * Jeannine M Petersen View author publications You can also search for this author inPubMed Google Scholar * Francois Balloux
View author publications You can also search for this author inPubMed Google Scholar * Paul Keim View author publications You can also search for this author inPubMed Google Scholar *
Thierry Wirth View author publications You can also search for this author inPubMed Google Scholar * Jacques Ravel View author publications You can also search for this author inPubMed
Google Scholar * Ruifu Yang View author publications You can also search for this author inPubMed Google Scholar * Elisabeth Carniel View author publications You can also search for this
author inPubMed Google Scholar * Mark Achtman View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS M.A., T.W., D.M.W., P.R., J.R., R.Y. and P.K.
designed the study. L.R., J.M.P., R.Y. and E.C. contributed _Y. pestis_ DNA and demographic information. G.M., Y.S., M.E., P.R., M.F., B.K., A.J.V., Y.L., Y.C., P.L. and N.R.T. performed
sequencing, SNP discovery, MassArray and SNP testing. G.M., Y.S., C.J.M., M.E., P.R., D.M.W. and P.L. performed bioinformatic analyses of the data. C.J.M., T.J., R.L., F.B. and T.W.
performed population genetic analyses. M.A., C.J.M., M.E., P.R., D.M.W., T.J., F.B., P.K., T.W., J.R., R.Y. and E.C. wrote the manuscript. CORRESPONDING AUTHORS Correspondence to Ruifu Yang,
Elisabeth Carniel or Mark Achtman. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY TEXT AND FIGURES
Supplementary Figures 1–5, Supplementary Tables 1 and 2 and Supplementary Note (PDF 3326 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Morelli, G.,
Song, Y., Mazzoni, C. _et al._ _Yersinia pestis_ genome sequencing identifies patterns of global phylogenetic diversity. _Nat Genet_ 42, 1140–1143 (2010). https://doi.org/10.1038/ng.705
Download citation * Received: 11 January 2010 * Accepted: 08 October 2010 * Published: 31 October 2010 * Issue Date: December 2010 * DOI: https://doi.org/10.1038/ng.705 SHARE THIS ARTICLE
Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided
by the Springer Nature SharedIt content-sharing initiative