Play all audios:
ABSTRACT Polycomb group (PcG) complexes are multiprotein assemblages that bind to chromatin and establish chromatin states leading to epigenetic silencing1,2. PcG proteins regulate homeotic
genes in flies and vertebrates, but little is known about other PcG targets and the role of the PcG in development, differentiation and disease. Here, we determined the distribution of the
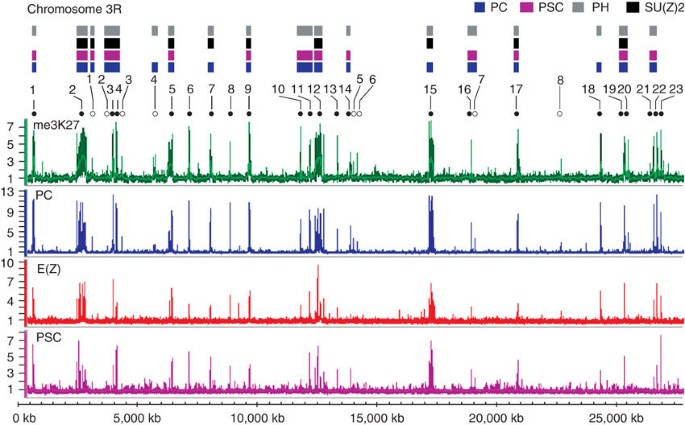
PcG proteins PC, E(Z) and PSC and of trimethylation of histone H3 Lys27 (me3K27) in the _D. melanogaster_ genome. At more than 200 PcG target genes, binding sites for the three PcG proteins
colocalize to presumptive Polycomb response elements (PREs). In contrast, H3 me3K27 forms broad domains including the entire transcription unit and regulatory regions. PcG targets are highly
enriched in genes encoding transcription factors, but they also include genes coding for receptors, signaling proteins, morphogens and regulators representing all major developmental
pathways. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to
this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy
now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer
support SIMILAR CONTENT BEING VIEWED BY OTHERS THE ROLES OF POLYCOMB REPRESSIVE COMPLEXES IN MAMMALIAN DEVELOPMENT AND CANCER Article 15 March 2021 ADAPTATION OF GENE LOCI TO
HETEROCHROMATIN IN THE COURSE OF _DROSOPHILA_ EVOLUTION IS ASSOCIATED WITH INSULATOR PROTEINS Article Open access 17 July 2020 TRANSIENT LOSS OF POLYCOMB COMPONENTS INDUCES AN EPIGENETIC
CANCER FATE Article Open access 24 April 2024 REFERENCES * Pirrotta, V. PcG complexes and chromatin silencing. _Curr. Opin. Genet. Dev._ 7, 249–258 (1997). Article CAS PubMed Google
Scholar * Lund, A.H. & van Lohuizen, M. Polycomb complexes and silencing mechanisms. _Curr. Opin. Cell Biol._ 16, 239–246 (2004). Article CAS PubMed Google Scholar * Rastelli, L.,
Chan, C.S. & Pirrotta, V. Related chromosome binding sites for zeste, suppressors of zeste and Polycomb group proteins in _Drosophila_ and their dependence on Enhancer of zeste function.
_EMBO J._ 12, 1513–1522 (1993). Article CAS PubMed PubMed Central Google Scholar * Chan, C.-S., Rastelli, L. & Pirrotta, V. A Polycomb response element in the _Ubx_ gene that
determines an epigenetically inherited state of repression. _EMBO J._ 13, 2553–2564 (1994). Article CAS PubMed PubMed Central Google Scholar * Sengupta, A.K., Kuhrs, A. & Muller, J.
General transcriptional silencing by a Polycomb response element in _Drosophila_. _Development_ 131, 1959–1965 (2004). Article CAS PubMed Google Scholar * Saurin, A.J., Shao, Z.,
Erdjument-Bromage, H., Tempst, P. & Kingston, R.E. A _Drosophila_ Polycomb group complex includes Zeste and dTAFII proteins. _Nature_ 412, 655–660 (2001). Article CAS PubMed Google
Scholar * Czermin, B. et al. _Drosophila_ Enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. _Cell_ 111, 185–196 (2002).
Article CAS PubMed Google Scholar * Cao, R. et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. _Science_ 298, 1039–1043 (2002). Article CAS PubMed Google
Scholar * Kuzmichev, A., Nishioka, K., Erdjument-Bromage, H., Tempst, P. & Reinberg, D. Histone methyltransferase activity associated with a human multiprotein complex containing the
Enhancer of Zeste protein. _Genes Dev._ 22, 2893–2905 (2002). Article Google Scholar * Müller, J. et al. Histone methyltransferase activity of a _Drosophila_ Polycomb Group repressor
complex. _Cell_ 111, 197–208 (2002). Article PubMed Google Scholar * Fischle, W. et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb
and HP1 chromodomains. _Genes Dev._ 17, 1870–1881 (2003). Article CAS PubMed PubMed Central Google Scholar * Jones, R.S. & Gelbart, W.M. Genetic analysis of the _Enhancer of zeste_
locus and its role in gene regulation in _Drosophila melanogaster_. _Genetics_ 126, 185–199 (1990). CAS PubMed PubMed Central Google Scholar * Phillips, M.D. & Shearn, A. Mutations
in _polycombeotic_, a _Drosophila_ Polycomb-group gene, cause a wide range of maternal and zygotic phenotypes. _Genetics_ 125, 91–101 (1990). CAS PubMed PubMed Central Google Scholar *
Strutt, H. & Paro, R. The polycomb group protein complex of _Drosophila melanogaster_ has different composition at different target genes. _Mol. Cell. Biol._ 17, 6773–6783 (1997).
Article CAS PubMed PubMed Central Google Scholar * Bloyer, S., Cavalli, G., Brock, H.W. & Dura, J.-M. Identification and characterization of polyhomeotic PREs and TREs. _Dev. Biol._
261, 426–442 (2003). Article CAS PubMed Google Scholar * Reim, I., Lee, H.-H. & Frasch, M. The T-box-encoding Dorsocross genes function in amnioserosa development and the patterning
of the dorsolateral germ band downstream of Dpp. _Development_ 130, 3187–3204 (2003). Article CAS PubMed Google Scholar * Jagla, K., Bellard, M. & Frasch, M. A cluster of
_Drosophila_ homeobox genes involved in mesoderm differentiation programs. _Bioessays_ 23, 125–133 (2001). Article CAS PubMed Google Scholar * Chiang, A., O'Connor, M.B., Paro, R.,
Simon, J. & Bender, W. Discrete Polycomb-binding sites in each parasegmental domain of the bithorax complex. _Development_ 121, 1681–1689 (1995). CAS PubMed Google Scholar * Shimell,
M.J., Peterson, A.J., Burr, J., Simon, J.A. & O'Connor, M. Functional analysis of repressor binding sites in the _iab-2_ regulatory region of the _abdominal-A_ homeotic gene. _Dev.
Biol._ 218, 38–52 (2000). Article CAS PubMed Google Scholar * Busturia, A., Wightman, C.D. & Sakonju, S. A silencer is required for maintenance of transcriptional repression
throughout _Drosophila_ development. _Development_ 124, 4343–4350 (1997). CAS PubMed Google Scholar * Mihaly, J., Hogga, I., Gausz, J., Gyurkovics, H. & Karch, F. In situ dissection
of the _Fab-7_ region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. _Development_ 124, 1809–1820 (1997). CAS PubMed Google Scholar * Barges, S.
et al. The _Fab-8_ boundary defines the distal limit of the bithorax complex _iab-7_ domain and insulates _iab-7_ from initiation elements and a PRE in the adjacent _iab-8_ domain.
_Development_ 127, 779–790 (2000). CAS PubMed Google Scholar * Orlando, V. & Paro, R. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde
cross-linked chromatin. _Cell_ 75, 1187–1198 (1993). Article CAS PubMed Google Scholar * Gene Ontology Consortium. The Gene Ontology (GO) database and informatics resource. _Nucleic
Acids Res._ 32, D258–D261 (2004). * Ringrose, L., Rehmsmeier, M., Dura, J.M. & Paro, R. Genome-wide prediction of Polycomb/Trithorax response elements in _Drosophila melanogaster_. _Dev.
Cell_ 5, 759–771 (2003). Article CAS PubMed Google Scholar * Brunk, B.P., Martin, E.C. & Adler, P.N. _Drosophila_ genes _Posterior Sex Combs_ and _Suppressor two of zeste_ encode
proteins with homology to the murine bmi-1 oncogene. _Nature_ 353, 351–353 (1991). Article CAS PubMed Google Scholar * Lee, T.I. et al. Control of developmental regulators by Polycomb in
human embryonic stem cells. _Cell_ 125, 301–313 (2006). Article CAS PubMed PubMed Central Google Scholar * Bernstein, B.E. et al. A bivalent chromatin structure marks key developmental
genes in embryonic stem cells. _Cell_ 125, 315–326 (2006). Article CAS PubMed Google Scholar * Négre, N. et al. Chromosomal distribution of PcG proteins during _Drosophila_ development.
_PLoS Biol._ 4, e170 (2006). Article PubMed PubMed Central Google Scholar * Schneider, I. Cell lines derived from the late embryonic stages of _Drosophila melanogaster_. _J. Embryol.
Exp. Morphol._ 27, 353–356 (1972). CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS We are grateful to T. Jenuwein for the anti-H3 me3K27 antibody; to D. McCabe for polytene
chromosome preparations; to A. Brooks, Q. Wang and V. Patel of the Bionomics Research and Technology Center of the Rutgers Environmental and Occupational Health Sciences Institute for
hybridization and scanning of the microarrays. Particular thanks to M. Eisen for leading the Berkeley Drosophila Transcription Network Project's development of ChIP/chip data analysis
methods and for encouraging this work. Work conducted by the BDTNP is funded by a grant from the US National Institute of General Medical Sciences and the US National Human Genome Research
Institute (GM704403) at Lawrence Berkeley National Laboratory under Department of Energy contract DE-AC02-05CH11231. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Molecular
Biology and Biochemistry, Rutgers University, 604 Allison Road, Piscataway, 08854, New Jersey, USA Yuri B Schwartz, Tatyana G Kahn & Vincenzo Pirrotta * Genomics Division, Berkeley
Drosophila Transcription Network Project, Lawrence Berkeley National Laboratory, Berkeley, 94720, California, USA David A Nix, Xiao-Yong Li & Mark Biggin * Department of Statistics,
University of California, Berkeley, Berkeley, 94720, California, USA Richard Bourgon * Affymetrix, 6550 Vallejo Street, Emeryville, 94608, California, USA David A Nix Authors * Yuri B
Schwartz View author publications You can also search for this author inPubMed Google Scholar * Tatyana G Kahn View author publications You can also search for this author inPubMed Google
Scholar * David A Nix View author publications You can also search for this author inPubMed Google Scholar * Xiao-Yong Li View author publications You can also search for this author
inPubMed Google Scholar * Richard Bourgon View author publications You can also search for this author inPubMed Google Scholar * Mark Biggin View author publications You can also search for
this author inPubMed Google Scholar * Vincenzo Pirrotta View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Vincenzo
Pirrotta. ETHICS DECLARATIONS COMPETING INTERESTS D.A.N. is employed by Affymetrix. SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIG. 1 Distribution of PcG proteins and H3 me3K27 mark along the X
chromosome. (PDF 81 kb) SUPPLEMENTARY FIG. 2 Distribution of PcG proteins and H3 me3K27 mark along chromosome 4. (PDF 84 kb) SUPPLEMENTARY FIG. 3 Distribution of PcG proteins and H3 me3K27
mark along 2L chromosome. (PDF 83 kb) SUPPLEMENTARY FIG. 4 Distribution of PcG proteins and H3 me3K27 mark along 2R chromosome. (PDF 81 kb) SUPPLEMENTARY FIG. 5 Distribution of PcG proteins
and H3 me3K27 mark along 3L chromosome. (PDF 82 kb) SUPPLEMENTARY FIG. 6 Co-localization between the binding of PSC and other PcG proteins. (PDF 44 kb) SUPPLEMENTARY FIG. 7 Co-localization
between the binding of E(Z) and other PcG proteins. (PDF 42 kb) SUPPLEMENTARY FIG. 8 Comparison of microarray data to the Ringrose _et al_. genome-wide prediction of PREs. (PDF 21 kb)
SUPPLEMENTARY FIG. 9 PcG proteins bind to _ph_ and _Psc-Su(z)2_ polycomb group genes. (PDF 129 kb) SUPPLEMENTARY FIG. 10 Valication of microarray hybridization results by real-time PCR. (PDF
207 kb) SUPPLEMENTARY TABLE 1 List of PcG sites. (PDF 196 kb) SUPPLEMENTARY TABLE 2 General features of PcG sites. (PDF 53 kb) SUPPLEMENTARY TABLE 3 List of PCR primers. (PDF 69 kb)
SUPPLEMENTARY METHODS (PDF 62 KB) SUPPLEMENTARY NOTE (PDF 12 KB) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Schwartz, Y., Kahn, T., Nix, D. _et al._
Genome-wide analysis of Polycomb targets in _Drosophila melanogaster_. _Nat Genet_ 38, 700–705 (2006). https://doi.org/10.1038/ng1817 Download citation * Received: 06 February 2006 *
Accepted: 01 May 2006 * Published: 28 May 2006 * Issue Date: 01 June 2006 * DOI: https://doi.org/10.1038/ng1817 SHARE THIS ARTICLE Anyone you share the following link with will be able to
read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative