Play all audios:
ABSTRACT The mammalian cell nucleus is a dynamic and highly organized structure. Most proteins are mobile within the nuclear compartment, and this mobility reflects transient interactions
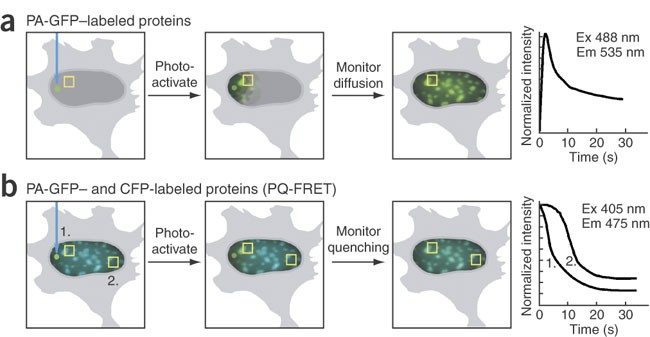
with chromatin, as well as network interactions with a variety of protein partners. To study these dynamic processes in living cells, we developed an imaging method that combines the
photoactivated green fluorescent protein (PA-GFP) and fluorescence resonance energy transfer (FRET) microscopy. We used this new method, photoquenching FRET (PQ-FRET), to define the dynamic
interactions of the heterochromatin protein-1 alpha (HP1α) and the transcription factor CCAAT/enhancer binding protein alpha (C/EBPα) in regions of centromeric heterochromatin in mouse
pituitary cells. The advantage of the PQ-FRET assay is that it provides simultaneous measurement of a protein's mobility, its exchange within macromolecular complexes and its
interactions with other proteins in the living cell without the need for corrections based on reference images acquired from control cells. Access through your institution Buy or subscribe
This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access
$259.00 per year only $21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are
calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS
PROXIMAL MOLECULAR PROBE TRANSFER (PROMPT), A NEW APPROACH FOR IDENTIFYING SITES OF PROTEIN/NUCLEIC ACID INTERACTION IN CELLS BY CORRELATED LIGHT AND ELECTRON MICROSCOPY Article Open access
05 December 2023 TAGBIFC TECHNIQUE ALLOWS LONG-TERM SINGLE-MOLECULE TRACKING OF PROTEIN-PROTEIN INTERACTIONS IN LIVING CELLS Article Open access 19 March 2021 EXTENDING FLUORESCENCE
ANISOTROPY TO LARGE COMPLEXES USING REVERSIBLY SWITCHABLE PROTEINS Article Open access 10 October 2022 REFERENCES * Lamond, A.I. & Earnshaw, W.C. Structure and function in the nucleus.
_Science_ 280, 547–553 (1998). Article CAS Google Scholar * Cremer, T. & Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. _Nat. Rev.
Genet._ 2, 292–301 (2001). Article CAS Google Scholar * Misteli, T. Concepts in nuclear architecture. _Bioessays_ 27, 477–487 (2005). Article CAS Google Scholar * Francastel, C.,
Magis, W. & Groudine, M. Nuclear relocation of a transactivator subunit precedes target gene activation. _Proc. Natl. Acad. Sci. USA_ 98, 12120–12125 (2001). Article CAS Google Scholar
* Remenyi, A., Scholer, H.R. & Wilmanns, M. Combinatorial control of gene expression. _Nat. Struct. Mol. Biol._ 11, 812–815 (2004). Article CAS Google Scholar * Amirand, C. et al.
Three distinct sub-nuclear populations of HMG-I protein of different properties revealed by co-localization image analysis. _J. Cell Sci._ 111, 3551–3561 (1998). CAS PubMed Google Scholar
* Perrod, S. & Gasser, S.M. Long-range silencing and position effects at telomeres and centromeres: parallels and differences. _Cell. Mol. Life Sci._ 60, 2303–2318 (2003). Article CAS
Google Scholar * Ficz, G., Heintzmann, R. & Arndt-Jovin, D.J. Polycomb group protein complexes exchange rapidly in living _Drosophila_. _Development_ 132, 3963–3976 (2005). Article
CAS Google Scholar * Brown, K.E. et al. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. _Cell_ 91, 845–854 (1997). Article CAS Google
Scholar * Francastel, C., Schübeler, D., Martin, D.I. & Groudine, M. Nuclear compartmentalization and gene activity. _Nat. Rev. Mol. Cell Biol._ 1, 137–143 (2000). Article CAS Google
Scholar * Piwien Pilipuk, G., Galigniana, M.D. & Schwartz, J. Subnuclear localization of C/EBP beta is regulated by growth hormone and dependent on MAPK. _J. Biol. Chem._ 278,
35668–35677 (2003). Article Google Scholar * Darlington, G.J., Wang, N. & Hanson, R.W. C/EBP alpha: a critical regulator of genes governing integrative metabolic processes. _Curr.
Opin. Genet. Dev._ 5, 565–570 (1995). Article CAS Google Scholar * Tang, Q.Q. & Lane, M.D. Activation and centromeric localization of CCAAT/enhancer-binding proteins during the
mitotic clonal expansion of adipocyte differentiation. _Genes Dev._ 13, 2231–2241 (1999). Article CAS Google Scholar * Schaufele, F. et al. CCAAT/enhancer binding protein alpha assembles
essential cooperating factors in common subnuclear domains. _Mol. Endocrinol._ 15, 1665–1676 (2001). CAS PubMed Google Scholar * Liu, W., Enwright, J.F., Hyun, W., Day, R.N. &
Schaufele, F. CCAAT/Enhancer Binding Protein alpha uses distinct domains to prolong pituitary cells in the Growth 1 and DNA Synthesis phases of the cell cycle. _BMC Cell Biol._ 3, 6 (2002).
Article Google Scholar * Cobb, B.S. et al. Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding. _Genes Dev._ 14, 2146–2160 (2000). Article CAS Google Scholar *
Tang, Q.Q. & Lane, M.D. Role of C/EBP homologous protein (CHOP-10) in the programmed activation of CCAAT/enhancer-binding protein-beta during adipogenesis. _Proc. Natl. Acad. Sci. USA_
97, 12446–12450 (2000). Article CAS Google Scholar * Patterson, G.H. & Lippincott-Schwartz, J. A photoactivatable GFP for selective photolabeling of proteins and cells. _Science_ 297,
1873–1877 (2002). Article CAS Google Scholar * Lippincott-Schwartz, J. & Patterson, G.H. Development and use of fluorescent protein markers in living cells. _Science_ 300, 87–91
(2003). Article CAS Google Scholar * Jares-Erijman, E.A. & Jovin, T.M. FRET imaging. _Nat. Biotechnol._ 21, 1387–1395 (2003). Article CAS Google Scholar * Day, R.N. &
Schaufele, F. Imaging molecular interactions in living cells. _Mol. Endocrinol._ 19, 1675–1686 (2005). Article CAS Google Scholar * Berney, C. & Danuser, G. FRET or no FRET: a
quantitative comparison. _Biophys. J._ 84, 3992–4010 (2003). Article CAS Google Scholar * Bastiaens, P.I. & Jovin, T.M. Microspectroscopic imaging tracks the intracellular processing
of a signal transduction protein: fluorescent-labeled protein kinase C beta I. _Proc. Natl. Acad. Sci. USA_ 93, 8407–8412 (1996). Article CAS Google Scholar * Maul, G.G., Negorev, D.,
Bell, P. & Ishov, A.M. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. _J. Struct. Biol._ 129, 278–287 (2000). Article CAS Google Scholar * Cheutin, T. et al.
Maintenance of stable heterochromatin domains by dynamic HP1 binding. _Science_ 299, 721–725 (2003). Article CAS Google Scholar * Krylov, D., Olive, M. & Vinson, C. Extending
dimerization interfaces: the bZIP basic region can form a coiled coil. _EMBO J._ 14, 5329–5337 (1995). Article CAS Google Scholar * Phair, R.D. et al. Global nature of dynamic
protein-chromatin interactions _in vivo:_ three dimensional genome scanning and dynamic interaction networks of chromatin proteins. _Mol. Cell. Biol._ 24, 6393–6402 (2004). Article CAS
Google Scholar * Maison, C. & Almouzni, G. HP1 and the dynamics of heterochromatin maintenance. _Nat. Rev. Mol. Cell Biol._ 5, 296–304 (2004). Article CAS Google Scholar * Bannister,
A.J. et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromodomain. _Nature_ 410, 120–124 (2001). Article CAS Google Scholar * Lukyanov, K.A., Chudakov, D.M.,
Lukyanov, S. & Verkhusha, V.V. Innovation: Photoactivatable fluorescent proteins. _Nat. Rev. Mol. Cell Biol._ 6, 885–891 (2005). Article CAS Google Scholar Download references
ACKNOWLEDGEMENTS We thank G. Patterson and J. Lippincott-Schwartz for kindly providing the PA-GFP–C1 vector, and R. Tsien for the mRFP1 cDNA. We thank M. Logsdon for technical assistance, Y.
Chen from the Keck Center for Cellular Imaging for assistance with FLIM data analysis, and F. Koberling (PicoQuant GmbH) for helpful discussion. We also thank J. Redick and C. Davis from
the Advanced Microscopy Facility for help with the laser scanning confocal microscopy. This work was supported by a grant from the US National Institutes of Health (DK47301 to R.N.D.).
AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Departments of Medicine and Cell Biology, P.O. Box 800578, University of Virginia Health Sciences Center, Charlottesville, 22908, Virginia, USA
Ignacio A Demarco, Cynthia F Booker & Richard N Day * W.M. Keck Center for Cellular Imaging, University of Virginia, Gilmer Hall, Charlottesville, 22904, Virginia, USA Ammasi Periasamy
Authors * Ignacio A Demarco View author publications You can also search for this author inPubMed Google Scholar * Ammasi Periasamy View author publications You can also search for this
author inPubMed Google Scholar * Cynthia F Booker View author publications You can also search for this author inPubMed Google Scholar * Richard N Day View author publications You can also
search for this author inPubMed Google Scholar CONTRIBUTIONS I.A.D. and R.N.D. contributed equally to the conceptual development of and implementation of this method. C.F.B. contributed to
all technical aspects of this project. A.P. developed and helped apply the fluorescence lifetime measurements. CORRESPONDING AUTHOR Correspondence to Richard N Day. ETHICS DECLARATIONS
COMPETING INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIG. 1 Wide-field microscope images showing the nuclei of mouse GHFT1 cells
that expressed either YFP-HP1α or YFP-C/EBPα. (PDF 535 kb) SUPPLEMENTARY FIG. 2 The full blot from the co-immunoprecipitation analysis of the association of HP1α and C/EBPα. (PDF 156 kb)
SUPPLEMENTARY FIG. 3 Control experiments monitoring the photobleaching of CFP under conditions used for the photoactivation of PA-GFP. (PDF 405 kb) SUPPLEMENTARY FIG. 4 The mean donor
lifetime distributions obtained by two-component analysis of CFP fluorescence lifetime from cells expressing: CFP-C/EBPα; CFP-C/EBPα and PA-GFP-HP1α; CFP-C/EBP BZIP and PA-GFP-CEBP BZIP.
(PDF 926 kb) SUPPLEMENTARY METHODS (DOC 29 KB) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Demarco, I., Periasamy, A., Booker, C. _et al._ Monitoring
dynamic protein interactions with photoquenching FRET. _Nat Methods_ 3, 519–524 (2006). https://doi.org/10.1038/nmeth889 Download citation * Received: 21 April 2006 * Accepted: 15 May 2006
* Published: 21 June 2006 * Issue Date: July 2006 * DOI: https://doi.org/10.1038/nmeth889 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get
shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative