Play all audios:
ABSTRACT Hydrothermal vent systems host microbial communities among which several microorganisms have been considered endemic to this type of habitat. It is still unclear how these organisms
colonize geographically distant hydrothermal environments. Based on 16S rRNA gene sequences, we compare the bacterial communities of sixteen Atlantic hydrothermal vent samples with our own
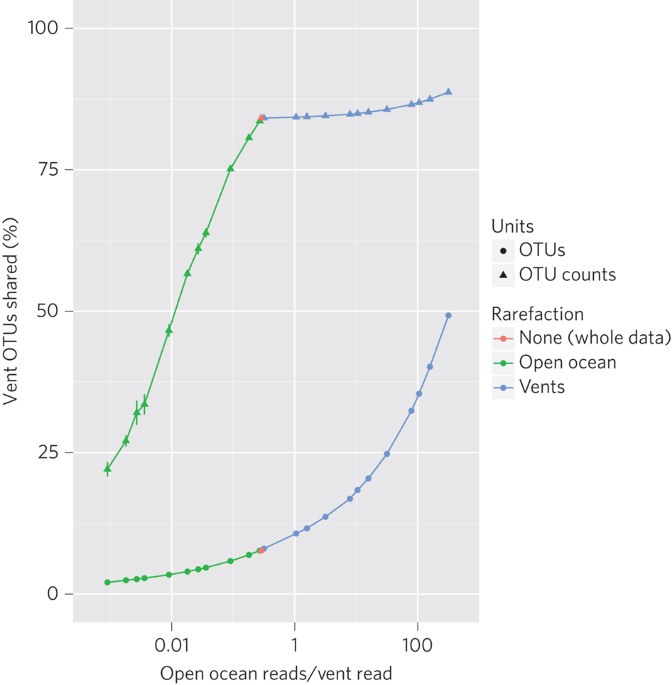
and publicly available global open ocean samples. Analysing sequences obtained from 63 million 16S rRNA genes, the genera we could identify in the open ocean waters contained 99.9% of the
vent reads. This suggests that previously observed vent exclusiveness is, in most cases, probably an artefact of lower sequencing depth. These findings are a further step towards elucidating
the role of the open ocean as a seed bank. They can explain the predicament of how species expected to be endemic to vent systems are able to colonize geographically distant hydrothermal
habitats and contribute to our understanding of whether ‘everything is really everywhere’. Access through your institution Buy or subscribe This is a preview of subscription content, access
via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 digital issues and online access to articles $119.00 per year only $9.92 per issue
Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL
ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS MICROBIAL COMMUNITIES OF AUKA
HYDROTHERMAL SEDIMENTS SHED LIGHT ON VENT BIOGEOGRAPHY AND THE EVOLUTIONARY HISTORY OF THERMOPHILY Article Open access 28 March 2022 A GENUS IN THE BACTERIAL PHYLUM AQUIFICOTA APPEARS TO BE
ENDEMIC TO AOTEAROA-NEW ZEALAND Article Open access 02 January 2024 NEW GLOBALLY DISTRIBUTED BACTERIAL PHYLA WITHIN THE FCB SUPERPHYLUM Article Open access 06 December 2022 REFERENCES *
Gibbons, S. M. _et al._ Evidence for a persistent microbial seed bank throughout the global ocean. _Proc. Natl Acad. Sci. USA_ 110, 4651–4655 (2013). Article Google Scholar * Campbell, B.
J., Engel, A. S., Porter, M. L. & Takai, K. The versatile _ε-proteobacteria_: key players in sulphidic habitats. _Nature Rev. Microbiol._ 4, 458–468 (2006). Article Google Scholar *
Sievert, S. M. & Vetriani, C. Chemoautotrophy at deep-sea vents: past, present, and future. _Oceanogr. Mar. Biol._ 25, 218–233 (2012). Google Scholar * Perner, M. _et al._ _In-situ_
chemistry and microbial community compositions in five deep-sea hydrothermal fluid samples from Irina II in the Logatchev field. _Environ. Microbiol._ 15, 1551–1560 (2013). Article Google
Scholar * Huber, J. A. _et al._ Isolated communities of _ε-proteobacteria_ in hydrothermal vent fluids of the Mariana Arc seamounts. _FEMS Microbiol. Ecol._ 73, 538–549 (2010). Google
Scholar * Beaulieu, S. E., Baker, E. T. & German, C. R. Where are the undiscovered hydrothermal vents on oceanic spreading ridges?. _Deep-Sea Res. II_ 121, 202–212 (2015). Article
Google Scholar * Stein, J. & Fisher, A. Observations and models of lateral hydrothermal circulation on a young ridge flank: numerical evaluation of thermal and chemical constraints.
_G__3_ 4, 1026 (2003). Google Scholar * Clarke, K. R. Non-parametric multivariate analyses of changes in community structure. _Aust. J. Ecol._ 18, 117–143 (1993). Article Google Scholar *
Sogin, M. L. _et al._ Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. _Proc. Natl Acad. Sci. USA_ 103, 12115–12120 (2006). Article Google Scholar * Galand, P.
E., Casamayor, E. O., Kirchman, D. L. & Lovejoy, C. Ecology of the rare microbial biosphere of the Arctic Ocean. _Proc. Natl Acad. Sci. USA_ 106, 22427–22432 (2009). Article Google
Scholar * Caporaso, J. G., Paszkiewicz, K., Field, D., Knight, R. & Gilbert, J. A. The western English Channel contains a persistent microbial seed bank. _ISME J._ 6, 1089–1093 (2012).
Article Google Scholar * Anderson, R., Sogin, M. L. & Baross, J. A. Biogeography and ecology of the rare and abundant microbial lineages in deep-sea hydrothermal vent. _FEMS Microbiol.
Ecol._ 91, 1–11 (2015). Article Google Scholar * Huber, J. A. _et al._ Microbial population structures in the deep marine biosphere. _Science_ 318, 97–100 (2007). Article Google Scholar
* Pedrós-Alió, C. Marine microbial diversity: can it be determined? _Trends Microbiol._ 14, 257–263 (2006). Article Google Scholar * Friedline, C., Franklin, R., McCallister, S. &
Rivera, M. Bacterial assemblages of the eastern Atlantic Ocean reveal both vertical and latitudinal biogeographic signatures. _Biogeosciences_ 9, 2177–2193 (2012). Article Google Scholar *
Sunagawa, S. _et al._ Ocean plankton. Structure and function of the global ocean microbiome. _Science_ 348, 1261359 (2015). Article Google Scholar * Rideout, J. R. _et al._ Subsampled
open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. _PeerJ_ 2, e545 (2014). Article Google Scholar * Baas Becking, L.
Geobiologie of Inleiding Tot de Milieukunde. in _Geobiology or Introduction to the Science of the Environment_ (Van Stockum & Zoon, 1934). Google Scholar * Liu, Z., Lozupone, C.,
Hamady, M., Bushman, F. D. & Knight, R. Short pyrosequencing reads suffice for accurate microbial community analysis. _Nucleic Acids Res._ 35, e120 (2007). Article Google Scholar *
Dworkin, M. & Falkow, S. _The Prokaryotes: A Handbook on the Biology of Bacteria_ 3rd edn (Springer, 2006). Book Google Scholar * Zielinski, F. U. _et al._ Widespread occurrence of an
intranuclear bacterial parasite in vent and seep bathymodiolin mussels. _Environ. Microbiol._ 11, 1150–1167 (2009). Article Google Scholar * Wentrup, C., Wendeberg, A., Schimak, M.,
Borowski, C. & Dubilier, N. Forever competent: deep-sea bivalves are colonized by their chemosynthetic symbionts throughout their lifetime. _Environ. Microbiol._ 16, 3699–3713 (2014).
Article Google Scholar * Rocap, G. _et al._ Genome divergence in two _Prochlorococcus_ ecotypes reflects oceanic niche differentiation. _Nature_ 424, 1042–1047 (2003). Article Google
Scholar * Perner, M. _et al._ Linking geology, fluid chemistry and microbial activity of basalt- and ultramafic-hosted deep-sea hydrothermal vent environments. _Geobiology_ 11, 340–355
(2013). Article Google Scholar * Garbe-Schönberg, D., Jähmlich, H., Koschinsky, A., Ratmeyer, V. & Westernströer, U. KIPS—a new multiport valve-based all-Teflon fluid sampling system
for ROVs. _EGU Meeting, Geophys. Res. Abstr._ 8, 07032 (2006). * Perner, M. _et al._ Short-term microbial and physico-chemical variability in low-temperature hydrothermal fluids near 5°S on
the Mid-Atlantic Ridge. _Environ. Microbiol._ 11, 2526–2541 (2009). Article Google Scholar * Klindworth, A. _et al._ Evaluation of general 16S ribosomal RNA gene PCR primers for classical
and next-generation sequencing-based diversity studies. _Nucleic Acids Res._ 41, e1 (2013). Article Google Scholar * Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible
trimmer for Illumina sequence data. _Bioinformatics_ 30, 2114–2120 (2014). Article Google Scholar * Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. _Nature
Methods_ 9, 357–359 (2012). Article Google Scholar * Magoc, T. & Salzberg, S. L. FLASH: fast length adjustment of short reads to improve genome assemblies. _Bioinformatics_ 27,
2957–2963 (2011). Article Google Scholar * Caporaso, J. G., Kuczynski, J. & Stombaugh, J. QIIME allows analysis of high-throughput community sequencing data. _Nature_ 7, 335–336
(2010). Google Scholar * Quast, C. _et al._ The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. _Nucleic Acids Res._ 41, D590–D596 (2013). Article
Google Scholar * Edgar, R. Search and clustering orders of magnitude faster than BLAST. _Bioinformatics_ 26, 2460–2461 (2010). Article Google Scholar * Caporaso, J. G. _et al._ PyNAST: a
flexible tool for aligning sequences to a template alignment. _Bioinformatics_ 26, 266–267 (2010). Article Google Scholar * Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree
2—approximately maximum-likelihood trees for large alignments. _PLoS ONE_ 5, e9490 (2010). Article Google Scholar * Chao, A. _et al._ Rarefaction and extrapolation with Hill numbers: a
framework for sampling and estimation in species diversity studies. _Ecol. Monogr._ 84, 45–67 (2014). Article Google Scholar * Lozupone, C., Lladser, M. E., Knights, D., Stombaugh, J.
& Knight, R. UniFrac: an effective distance metric for microbial community comparison. _ISME J._ 5, 169–172 (2011). Article Google Scholar * Leinonen, R., Sugawara, H. & Shumway,
M. The sequence read archive. _Nucleic Acids Res._ 39, D19–D21 (2011). Article Google Scholar * Huse, S. M. _et al._ VAMPS: a website for visualization and analysis of microbial population
structures. _BMC Bioinformatics_ 15, 41 (2014). Article Google Scholar * Egbert, G. & Erofeeva, S. Efficient inverse modeling of barotropic ocean tides. _J. Atmos. Ocean. Technol._
19, 183–204 (2002). Article Google Scholar * Walter, M. _et al._ Rapid dispersal of a hydrothermal plume by turbulent mixing. _Deep-Sea Res I_ 57, 931–945 (2010). Article Google Scholar
* Carpenter, J. H. The Chesapeake Bay Institute technique for the Winkler dissolved oxygen method. _Limnol. Oceanogr._ 10, 141–143 (1965). Article Google Scholar * Cline, J.
Spectrophotometric determination of hydrogen sulfide in natural waters. _Limnol. Oceanogr._ 14, 454–458 (1969). Article Google Scholar * Perner, M. _et al._ The influence of ultramafic
rocks on microbial communities at the Logatchev hydrothermal field, located 15°N on the Mid-Atlantic Ridge. _FEMS Microbiol. Ecol._ 61, 97–109 (2007). Article Google Scholar * Seifert, R.
_et al._ Ethylene and methane in the upper water column of the subtropical Atlantic. _Biogeochemistry_ 44, 73–91 (1999). Google Scholar * Schmidt, K. A. _et al._ Fluid elemental and stable
isotope composition of the Nibelungen hydrothermal field (8°18′S, Mid-Atlantic Ridge): constraints on fluid–rock interaction in heterogeneous lithosphere. _Chem. Geol._ 280, 1–18 (2011).
Article Google Scholar * Schmidt, K., Koschinsky, A., Garbe-Schönberg, D., de Carvalho, L. M. & Seifert, R. Geochemistry of hydrothermal fluids from the ultramafic-hosted Logatchev
hydrothermal field, 15°N on the Mid-Atlantic Ridge: temporal and spatial investigation. _Chem. Geol._ 242, 1–21 (2007). Article Google Scholar * Benjamini, Y. & Hochberg, Y.
Controlling the false discovery rate: a practical and powerful approach to multiple testing. _J. R. Stat. Soc. Ser. B_ 57, 289–300 (1995). Google Scholar Download references
ACKNOWLEDGEMENTS The authors thank the captain and crews of the research vessels and _ROV Kiel6000_ (GEOMAR, Kiel) for helping us to obtain deep-sea vent samples. The authors also thank H.
Strauss for providing data on hydrogen sulfide and M. Alawi for discussions regarding the sequencing and bioinformatics analysis. The work was supported by grants from priority programme
1144 ‘From Mantle to Ocean: Energy-, Material- and Life-cycles at Spreading Axes’ of the German Science Foundation (DFG). S.B. was funded by grant no. DFG PE 1549-6/1. AUTHOR INFORMATION
AUTHORS AND AFFILIATIONS * Center for Bioinformatics, University of Hamburg, Bundesstraße 43, 20146 Hamburg, Germany Giorgio Gonnella & Stefan Kurtz * Molecular Biology of Microbial
Consortia, University of Hamburg, Biocenter Klein Flottbek, Ohnhorststraße 18, 22609 Hamburg, Germany Stefanie Böhnke & Mirjam Perner * Heinrich-Pette-Institut, Leibniz Institute for
Experimental Virology, Martinistraße 52, 20251 Hamburg, Germany Daniela Indenbirken * Institute of Geosciences, University of Kiel, Ludewig-Meyn-Straße 10, 24118 Kiel, Germany Dieter
Garbe-Schönberg * Institute for Geology, University of Hamburg, Bundesstraße 55, 20146 Hamburg, Germany Richard Seifert * Institute of Environmental Physics, University Bremen,
Otto-Hahn-Allee 1, 28359 Bremen, Germany Christian Mertens Authors * Giorgio Gonnella View author publications You can also search for this author inPubMed Google Scholar * Stefanie Böhnke
View author publications You can also search for this author inPubMed Google Scholar * Daniela Indenbirken View author publications You can also search for this author inPubMed Google
Scholar * Dieter Garbe-Schönberg View author publications You can also search for this author inPubMed Google Scholar * Richard Seifert View author publications You can also search for this
author inPubMed Google Scholar * Christian Mertens View author publications You can also search for this author inPubMed Google Scholar * Stefan Kurtz View author publications You can also
search for this author inPubMed Google Scholar * Mirjam Perner View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS M.P. designed the research
project, planned the sequencing and wrote the paper. G.G. co-planned the sequencing, performed bioinformatics and statistical analyses and wrote the paper. S.B. performed experiments. D.I.
performed sequencing. D.G.-S. contributed fluid elemental compositions and geochemical data. R.S. measured hydrogen and methane concentrations. C.M. performed tidal measurements. S.K.
advised on the bioinformatics analyses and wrote the paper. CORRESPONDING AUTHOR Correspondence to Mirjam Perner. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing
financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION Supplementary Results and Discussion 1-4, Supplementary Figures 1-6, Supplementary Tables 1-4, Supplementary Table
legends 5-10, and Supplementary References (PDF 896 kb) SUPPLEMENTARY TABLE 5 Genera shared between hydrothermal vent and open ocean samples. (XLSX 190 kb) SUPPLEMENTARY TABLE 6
Open-reference based OTUs shared between hydrothermal vent and open ocean samples. (XLSX 1109 kb) SUPPLEMENTARY TABLE 7 Environmental parameters of the different samples used for correlation
analyses. (XLSX 14 kb) SUPPLEMENTARY TABLE 8 Spearman correlations for the different sites between environmental parameters and bacterial classes. (XLSX 31 kb) SUPPLEMENTARY TABLE 9
Spearman correlations for the different sites between environmental parameters and bacterial genera. (XLSX 78 kb) SUPPLEMENTARY TABLE 10 Spearman correlations for the different sites between
environmental parameters and bacterial OTUs. (XLSX 2668 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Gonnella, G., Böhnke, S., Indenbirken, D.
_et al._ Endemic hydrothermal vent species identified in the open ocean seed bank. _Nat Microbiol_ 1, 16086 (2016). https://doi.org/10.1038/nmicrobiol.2016.86 Download citation * Received:
05 November 2015 * Accepted: 03 May 2016 * Published: 13 June 2016 * DOI: https://doi.org/10.1038/nmicrobiol.2016.86 SHARE THIS ARTICLE Anyone you share the following link with will be able
to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative