Play all audios:
ABSTRACT Oligomeric complexes of Trax and Translin proteins, known as C3POs, participate in several eukaryotic nucleic acid metabolism pathways, including RNA interference and tRNA
processing. In RNA interference in humans and _Drosophila_, C3PO activates the RNA-induced silencing complex (RISC) by removing the passenger strand of the small interfering RNA precursor
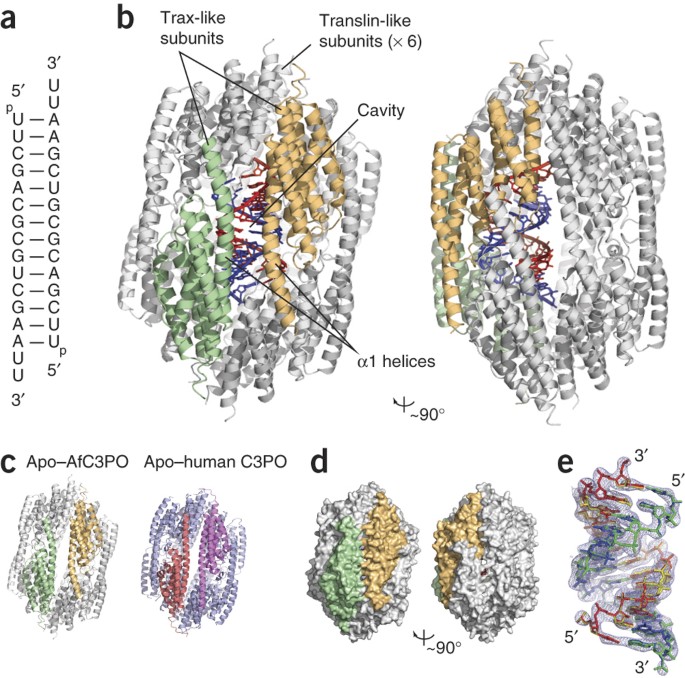
duplex, using nuclease activity present in Trax. How C3POs engage with nucleic acid substrates is unknown. Here we identify a single protein from _Archaeoglobus fulgidus_ that assembles into
an octamer highly similar to human C3PO. The structure in complex with duplex RNA reveals that the octamer entirely encapsulates a single 13-base-pair RNA duplex inside a large inner
cavity. Trax-like-subunit catalytic sites target opposite strands of the duplex for cleavage separated by 7 base pairs. The structure provides insight into the mechanism of RNA recognition
and cleavage by an archaeal C3PO-like complex. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access
through your institution Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink *
Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional
subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS DYNAMIC MECHANISMS OF CRISPR INTERFERENCE BY _ESCHERICHIA COLI_ CRISPR-CAS3 Article Open
access 30 August 2022 RPA TRANSFORMS RNASE H1 TO A BIDIRECTIONAL EXORIBONUCLEASE FOR PROCESSIVE RNA–DNA HYBRID CLEAVAGE Article Open access 29 August 2024 NUCLEIC-ACID-TRIGGERED NADASE
ACTIVATION OF A SHORT PROKARYOTIC ARGONAUTE Article 02 October 2023 ACCESSION CODES PRIMARY ACCESSIONS PROTEIN DATA BANK * 3ZC0 * 3ZC1 REFERENCED ACCESSIONS PROTEIN DATA BANK * 3PJA
REFERENCES * Ghildiyal, M. & Zamore, P.D. Small silencing RNAs: an expanding universe. _Nat. Rev. Genet._ 10, 94–108 (2009). CAS PubMed PubMed Central Google Scholar * Kim, V.N.,
Han, J. & Siomi, M.C. Biogenesis of small RNAs in animals. _Nat. Rev. Mol. Cell Biol._ 10, 126–139 (2009). CAS PubMed Google Scholar * Czech, B. & Hannon, G.J. Small RNA sorting:
matchmaking for Argonautes. _Nat. Rev. Genet._ 12, 19–31 (2011). CAS PubMed Google Scholar * Kawamata, T. & Tomari, Y. Making RISC. _Trends Biochem. Sci._ 35, 368–376 (2010). CAS
PubMed Google Scholar * Nykänen, A., Haley, B. & Zamore, P.D. ATP requirements and small interfering RNA structure in the RNA interference pathway. _Cell_ 107, 309–321 (2001). PubMed
Google Scholar * Tomari, Y. et al. RISC assembly defects in the _Drosophila_ RNAi mutant armitage. _Cell_ 116, 831–841 (2004). CAS PubMed Google Scholar * Pham, J.W., Pellino, J.L., Lee,
Y.S., Carthew, R.W. & Sontheimer, E.J.A. Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in _Drosophila_. _Cell_ 117, 83–94 (2004). CAS PubMed Google Scholar *
Kawamata, T., Seitz, H. & Tomari, Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. _Nat. Struct. Mol. Biol._ 16, 953–960 (2009). CAS PubMed
Google Scholar * Yoda, M. et al. ATP-dependent human RISC assembly pathways. _Nat. Struct. Mol. Biol._ 17, 17–23 (2010). CAS PubMed Google Scholar * Iwasaki, S. et al. Hsc70/Hsp90
chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. _Mol. Cell_ 39, 292–299 (2010). CAS PubMed Google Scholar * Miyoshi, T., Takeuchi, A., Siomi, H. &
Siomi, M.C. A direct role for Hsp90 in pre-RISC formation in _Drosophila_. _Nat. Struct. Mol. Biol._ 17, 1024–1026 (2010). CAS PubMed Google Scholar * Johnston, M., Geoffroy, M.C.,
Sobala, A., Hay, R. & Hutvagner, G. HSP90 protein stabilizes unloaded argonaute complexes and microscopic P-bodies in human cells. _Mol. Biol. Cell_ 21, 1462–1469 (2010). CAS PubMed
PubMed Central Google Scholar * Liu, Q. et al. R2D2, a bridge between the initiation and effector steps of the _Drosophila_ RNAi pathway. _Science_ 301, 1921–1925 (2003). CAS PubMed
Google Scholar * Lee, Y.S. et al. Distinct roles for _Drosophila_ Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. _Cell_ 117, 69–81 (2004). CAS PubMed Google Scholar * Liu,
X., Jiang, F., Kalidas, S., Smith, D. & Liu, Q. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. _RNA_ 12, 1514–1520 (2006). CAS PubMed PubMed
Central Google Scholar * Liu, Y. et al. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. _Science_ 325, 750–753 (2009). CAS PubMed PubMed Central Google
Scholar * Ye, X. et al. Structure of C3PO and mechanism of human RISC activation. _Nat. Struct. Mol. Biol._ 18, 650–657 (2011). CAS PubMed PubMed Central Google Scholar * Schwarz, D.S.
et al. Asymmetry in the assembly of the RNAi enzyme complex. _Cell_ 115, 199–208 (2003). CAS PubMed Google Scholar * Khvorova, A., Reynolds, A. & Jayasena, S.D. Functional siRNAs and
miRNAs exhibit strand bias. _Cell_ 115, 209–216 (2003). CAS PubMed Google Scholar * Miyoshi, K., Tsukumo, H., Nagami, T., Siomi, H. & Siomi, M.C. Slicer function of _Drosophila_
Argonautes and its involvement in RISC formation. _Genes Dev._ 19, 2837–2848 (2005). CAS PubMed PubMed Central Google Scholar * Rand, T.A., Petersen, S., Du, F. & Wang, X. Argonaute2
cleaves the anti-guide strand of siRNA during RISC activation. _Cell_ 123, 621–629 (2005). CAS PubMed Google Scholar * Matranga, C., Tomari, Y., Shin, C., Bartel, D.P. & Zamore, P.D.
Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. _Cell_ 123, 607–620 (2005). CAS PubMed Google Scholar * Leuschner, P.J., Ameres, S.L.,
Kueng, S. & Martinez, J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. _EMBO Rep._ 7, 314–320 (2006). CAS PubMed PubMed Central Google Scholar * Wang,
Y., Sheng, G., Juranek, S., Tuschl, T. & Patel, D.J. Structure of the guide-strand-containing argonaute silencing complex. _Nature_ 456, 209–213 (2008). CAS PubMed PubMed Central
Google Scholar * Schirle, N.T. & MacRae, I.J. The crystal structure of human Argonaute2. _Science_ 336, 1037–1040 (2012). CAS PubMed PubMed Central Google Scholar * Nakanishi, K.,
Weinberg, D.E., Bartel, D.P. & Patel, D.J. Structure of yeast Argonaute with guide RNA. _Nature_ 486, 368–374 (2012). CAS PubMed PubMed Central Google Scholar * Elkayam, E. et al.
The structure of human Argonaute-2 in complex with miR-20a. _Cell_ 150, 100–110 (2012). CAS PubMed PubMed Central Google Scholar * Jaendling, A. & McFarlane, R.J. Biological roles of
translin and translin-associated factor-X: RNA metabolism comes to the fore. _Biochem. J._ 429, 225–234 (2010). CAS PubMed Google Scholar * Li, L. et al. The translin-TRAX complex (C3PO)
is a ribonuclease in tRNA processing. _Nat. Struct. Mol. Biol._ 19, 824–830 (2012). CAS PubMed PubMed Central Google Scholar * Tian, Y. et al. Multimeric assembly and biochemical
characterization of the Trax-translin endonuclease complex. _Nat. Struct. Mol. Biol._ 18, 658–664 (2011). CAS PubMed PubMed Central Google Scholar * Pascal, J.M., Hart, P.J., Hecht, N.B.
& Robertus, J.D. Crystal structure of TB-RBP, a novel RNA-binding and regulating protein. _J. Mol. Biol._ 319, 1049–1057 (2002). CAS PubMed Google Scholar * Sugiura, I. et al.
Structure of human translin at 2.2 Å resolution. _Acta Crystallogr. D Biol. Crystallogr._ 60, 674–679 (2004). PubMed Google Scholar * Chennathukuzhi, V. et al. Mice deficient for
testis-brain RNA-binding protein exhibit a coordinate loss of TRAX, reduced fertility, altered gene expression in the brain, and behavioral changes. _Mol. Cell. Biol._ 23, 6419–6434 (2003).
CAS PubMed PubMed Central Google Scholar * Yang, S. et al. Translin-associated factor X is post-transcriptionally regulated by its partner protein TB-RBP, and both are essential for
normal cell proliferation. _J. Biol. Chem._ 279, 12605–12614 (2004). CAS PubMed Google Scholar * Gupta, G.D. et al. Co-expressed recombinant human Translin-Trax complex binds DNA. _FEBS
Lett._ 579, 3141–3146 (2005). CAS PubMed Google Scholar * Claussen, M., Koch, R., Jin, Z.Y. & Suter, B. Functional characterization of _Drosophila_ Translin and Trax. _Genetics_ 174,
1337–1347 (2006). CAS PubMed PubMed Central Google Scholar * Jaendling, A., Ramayah, S., Pryce, D.W. & McFarlane, R.J. Functional characterisation of the _Schizosaccharomyces pombe_
homologue of the leukaemia-associated translocation breakpoint binding protein translin and its binding partner, TRAX. _Biochim. Biophys. Acta_ 1783, 203–213 (2008). CAS PubMed Google
Scholar * Aoki, K., Suzuki, K., Ishida, R. & Kasai, M. The DNA binding activity of Translin is mediated by a basic region in the ring-shaped structure conserved in evolution. _FEBS
Lett._ 443, 363–366 (1999). CAS PubMed Google Scholar * Chennathukuzhi, V.M., Kurihara, Y., Bray, J.D. & Hecht, N.B. Trax (translin-associated factor X), a primarily cytoplasmic
protein, inhibits the binding of TB-RBP (translin) to RNA. _J. Biol. Chem._ 276, 13256–13263 (2001). CAS PubMed Google Scholar * Eliahoo, E. et al. Mapping of interaction sites of the
_Schizosaccharomyces pombe_ protein Translin with nucleic acids and proteins: a combined molecular genetics and bioinformatics study. _Nucleic Acids Res._ 38, 2975–2989 (2010). CAS PubMed
PubMed Central Google Scholar * Gupta, G.D. & Kumar, V. Identification of nucleic acid binding sites on translin-associated factor X (TRAX) protein. _PLoS ONE_ 7, e33035 (2012). CAS
PubMed PubMed Central Google Scholar * Heidenreich, O., Pieken, W. & Eckstein, F. Chemically modified RNA: approaches and applications. _FASEB J._ 7, 90–96 (1993). CAS PubMed Google
Scholar * Parker, J.S., Roe, S.M. & Barford, D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. _EMBO J._ 23, 4727–4737 (2004). CAS
PubMed PubMed Central Google Scholar * Winter, G. _xia2_: an expert system for macromolecular crystallography data reduction. _J. Appl. Crystallogr._ 43, 186–190 (2010). CAS Google
Scholar * Kabsch, W. XDS. _Acta Crystallogr. D Biol. Crystallogr._ 66, 125–132 (2010). CAS PubMed PubMed Central Google Scholar * Evans, P.R. An introduction to data reduction:
space-group determination, scaling and intensity statistics. _Acta Crystallogr. D Biol. Crystallogr._ 67, 282–292 (2011). CAS PubMed PubMed Central Google Scholar * Collaborative
Computational Project. N. The CCP4 suite: programs for protein crystallography. _Acta Crystallogr. D Biol. Crystallogr._ 50, 760–763 (1994). * Blessing, R.H. & Smith, G.D. Difference
structure-factor normalization for heavy-atom or anomalous-scattering substructure determinations. _J. Appl. Crystallogr._ 32, 664–670 (1999). CAS Google Scholar * Weeks, C.M. &
Miller, R. The design and implementation of _SnB_ v2.0. _J. Appl. Crystallogr._ 32, 120–124 (1999). CAS Google Scholar * Vonrhein, C., Blanc, E., Roversi, P. & Bricogne, G. Automated
structure solution with autoSHARP. _Methods Mol. Biol._ 364, 215–230 (2007). CAS PubMed Google Scholar * Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular
structure solution. _Acta Crystallogr. D Biol. Crystallogr._ 66, 213–221 (2010). CAS PubMed PubMed Central Google Scholar * Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features
and development of _Coot_. _Acta Crystallogr. D Biol. Crystallogr._ 66, 486–501 (2010). CAS PubMed PubMed Central Google Scholar * McCoy, A.J. et al. _Phaser_ crystallographic software.
_J. Appl. Crystallogr._ 40, 658–674 (2007). CAS PubMed PubMed Central Google Scholar * Bond, C.S. & Schuttelkopf, A.W. _ALINE_: a WYSIWYG protein-sequence alignment editor for
publication-quality alignments. _Acta Crystallogr. D Biol. Crystallogr._ 65, 510–512 (2009). CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS We thank staff at Diamond Light
Source, UK for help with data collection. This work was funded by a UK Medical Research Council Career Development Award to J.S.P. (grant no. G0600097). The pTwo-E vector was a gift from A.
Oliver, University of Sussex, Brighton, UK. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Biochemistry, University of Oxford, Oxford, UK Eneida A Parizotto, Edward D Lowe
& James S Parker Authors * Eneida A Parizotto View author publications You can also search for this author inPubMed Google Scholar * Edward D Lowe View author publications You can also
search for this author inPubMed Google Scholar * James S Parker View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS E.A.P. produced and
purified the proteins, grew the crystals and performed the biochemical assays. E.D.L. provided advice, maintained facilities and assisted with X-ray data collection. J.S.P. collected X-ray
data, determined the structures and wrote the manuscript. CORRESPONDING AUTHOR Correspondence to James S Parker. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing
financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY TEXT AND FIGURES Supplementary Figures 1–6 (PDF 3102 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE
THIS ARTICLE Parizotto, E., Lowe, E. & Parker, J. Structural basis for duplex RNA recognition and cleavage by _Archaeoglobus fulgidus_ C3PO. _Nat Struct Mol Biol_ 20, 380–386 (2013).
https://doi.org/10.1038/nsmb.2487 Download citation * Received: 20 September 2012 * Accepted: 10 December 2012 * Published: 27 January 2013 * Issue Date: March 2013 * DOI:
https://doi.org/10.1038/nsmb.2487 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently
available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative