Play all audios:
ABSTRACT The principal function of CRISPR–Cas systems in archaea and bacteria is defence against mobile genetic elements (MGEs), including viruses, plasmids and transposons. However, the
relationships between CRISPR–Cas and MGEs are far more complex. Several classes of MGE contributed to the origin and evolution of CRISPR–Cas, and, conversely, CRISPR–Cas systems and their
components were recruited by various MGEs for functions that remain largely uncharacterized. In this Analysis article, we investigate and substantially expand the range of CRISPR–Cas
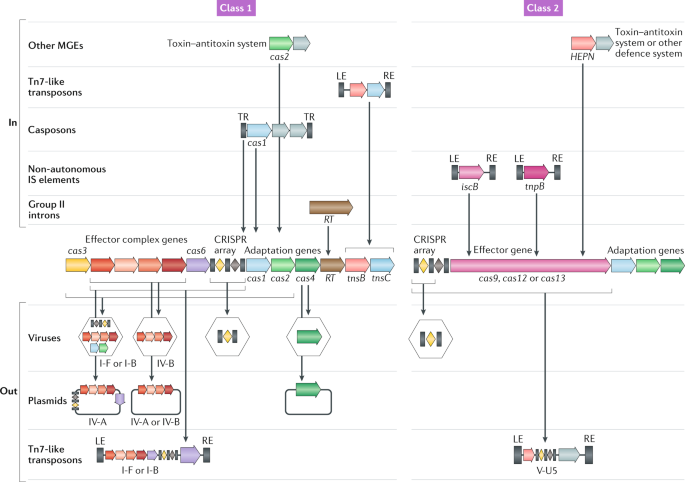
components carried by MGEs. Three groups of Tn7-like transposable elements encode ‘minimal’ type I CRISPR–Cas derivatives capable of target recognition but not cleavage, and another group
encodes an inactivated type V variant. These partially inactivated CRISPR–Cas variants might mediate guide RNA-dependent integration of the respective transposons. Numerous plasmids and some
prophages encode type IV systems, with similar predicted properties, that appear to contribute to competition among plasmids and between plasmids and viruses. Many prokaryotic viruses also
carry CRISPR mini-arrays, some of which recognize other viruses and are implicated in inter-virus conflicts, and solitary repeat units, which could inhibit host CRISPR–Cas systems. Access
through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other
Nature Portfolio journals Get Nature+, our best-value online-access subscription $32.99 / 30 days cancel any time Learn more Subscribe to this journal Receive 12 print issues and online
access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which
are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS
ALTERNATIVE FUNCTIONS OF CRISPR–CAS SYSTEMS IN THE EVOLUTIONARY ARMS RACE Article 06 January 2022 TRANSPOSON-ENCODED NUCLEASES USE GUIDE RNAS TO PROMOTE THEIR SELFISH SPREAD Article 27
September 2023 MECHANISTIC AND EVOLUTIONARY INSIGHTS INTO A TYPE V-M CRISPR–CAS EFFECTOR ENZYME Article Open access 17 July 2023 REFERENCES * Makarova, K. S. et al. An updated evolutionary
classification of CRISPR-Cas systems. _Nat. Rev. Microbiol._ 13, 722–736 (2015). Article CAS PubMed PubMed Central Google Scholar * Mohanraju, P. et al. Diverse evolutionary roots and
mechanistic variations of the CRISPR-Cas systems. _Science_ 353, aad5147 (2016). Article PubMed Google Scholar * Barrangou, R. & Horvath, P. A decade of discovery: CRISPR functions
and applications. _Nat. Microbiol._ 2, 17092 (2017). Article CAS PubMed Google Scholar * Jackson, S. A. et al. CRISPR-Cas: adapting to change. _Science_ 356, eaal5056 (2017). Article
PubMed Google Scholar * Koonin, E. V., Makarova, K. S. & Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. _Curr. Opin. Microbiol._ 37, 67–78 (2017). THIS
ARTICLE IS THE LATEST PUBLISHED OVERVIEW OF THE CRISPR–CAS DIVERSITY, WITH AN EMPHASIS ON CLASS 2 SYSTEMS DISCOVERED THROUGH DEDICATED SEARCH EFFORTS. Article CAS PubMed PubMed Central
Google Scholar * Garcia-Martinez, J., Maldonado, R. D., Guzman, N. M. & Mojica, F. J. M. The CRISPR conundrum: evolve and maybe die, or survive and risk stagnation. _Microb. Cell_ 5,
262–268 (2018). Article CAS PubMed PubMed Central Google Scholar * Makarova, K. S., Wolf, Y. I. & Koonin, E. V. Classification and nomenclature of CRISPR-Cas systems: where from
here? _CRISPR J_. https://doi.org/10.1089/crispr.2018.0033 (2018). Article PubMed Google Scholar * Faure, G., Makarova, K. S. & Koonin, E. V. CRISPR-Cas: complex functional networks
and multiple roles beyond adaptive immunity. _J. Mol. Biol._ 431, 3–20 (2018). Article PubMed Google Scholar * Westra, E. R., Buckling, A. & Fineran, P. C. CRISPR-Cas systems: beyond
adaptive immunity. _Nat. Rev. Microbiol._ 12, 317–326 (2014). Article CAS PubMed Google Scholar * Shmakov, S. A., Makarova, K. S., Wolf, Y. I., Severinov, K. V. & Koonin, E. V.
Systematic prediction of genes functionally linked to CRISPR-Cas systems by gene neighborhood analysis. _Proc. Natl Acad. Sci. USA_ 115, E5307–E5316 (2018). THIS ARTICLE PRESENTS SYSTEMATIC
PREDICTION AND ANALYSIS OF GENES ASSOCIATED WITH VARIOUS SUBSETS OF CRISPR–CAS SYSTEMS. THE RESULTS SUGGEST SUBSTANTIAL FUNCTIONAL DIVERSIFICATION OF CRISPR–CAS, IN PARTICULAR, COUPLING WITH
SIGNAL TRANSDUCTION, ESPECIALLY IN TYPE III SYSTEMS. Article CAS PubMed Google Scholar * Shah, S. A. et al. Comprehensive search for accessory proteins encoded with archaeal and
bacterial type III CRISPR-cas gene cassettes reveals 39 new cas gene families. _RNA Biol_. https://doi.org/10.1080/15476286.2018.1483685 (2018). THIS PAPER COMPLEMENTS SHMAKOV ET AL. (2018)
BY PROVIDING SYSTEMATIC ANALYSIS OF PREDICTED ACCESSORY PROTEINS ASSOCIATED WITH TYPE III CRISPR–CAS SYSTEMS. Article PubMed PubMed Central Google Scholar * Koonin, E. V. & Makarova,
K. S. Mobile genetic elements and evolution of CRISPR-Cas systems: all the way there and back. _Genome Biol. Evol_ 9, 2812–2825 (2017). Article CAS PubMed PubMed Central Google Scholar
* Krupovic, M., Beguin, P. & Koonin, E. V. Casposons: mobile genetic elements that gave rise to the CRISPR-Cas adaptation machinery. _Curr. Opin. Microbiol._ 38, 36–43 (2017). Article
CAS PubMed PubMed Central Google Scholar * Krupovic, M., Makarova, K. S., Forterre, P., Prangishvili, D. & Koonin, E. V. Casposons: a new superfamily of self-synthesizing DNA
transposons at the origin of prokaryotic CRISPR-Cas immunity. _BMC Biol._ 12, 36 (2014). Article PubMed PubMed Central Google Scholar * Kazazian, H. H. Jr. Mobile elements: drivers of
genome evolution. _Science_ 303, 1626–1632 (2004). Article CAS PubMed Google Scholar * Gueguen, E., Rousseau, P., Duval-Valentin, G. & Chandler, M. The transpososome: control of
transposition at the level of catalysis. _Trends Microbiol._ 13, 543–549 (2005). Article CAS PubMed Google Scholar * Peters, J. E. & Craig, N. L. Tn7: smarter than we thought. _Nat.
Rev. Mol. Cell Biol._ 2, 806–814 (2001). Article CAS PubMed Google Scholar * Fricker, A. D. & Peters, J. E. Vulnerabilities on the lagging-strand template: opportunities for mobile
elements. _Annu. Rev. Genet._ 48, 167–186 (2014). Article CAS PubMed Google Scholar * Nunez, J. K., Lee, A. S., Engelman, A. & Doudna, J. A. Integrase-mediated spacer acquisition
during CRISPR-Cas adaptive immunity. _Nature_ 519, 193–198 (2015). Article CAS PubMed PubMed Central Google Scholar * Hudaiberdiev, S. et al. Phylogenomics of Cas4 family nucleases.
_BMC Evol. Biol._ 17, 232 (2017). Article PubMed PubMed Central Google Scholar * Shmakov, S. et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. _Mol.
Cell_ 60, 385–397 (2015). Article CAS PubMed PubMed Central Google Scholar * Shmakov, S. et al. Diversity and evolution of class 2 CRISPR-Cas systems. _Nat. Rev. Microbiol._ 15,
169–182 (2017). THIS ARTICLE PRESENTS A DEFINITIVE DESCRIPTION OF THE DEDICATED EFFORTS ON THE DISCOVERY OF DIVERSE CLASS 2 CRISPR–CAS SYSTEMS. THE KEY FINDING IS THE IDENTIFICATION OF
MULTIPLE VARIANTS ASSIGNED TO SUBTYPE V-U THAT APPEAR TO HAVE INDEPENDENTLY EVOLVED FROM DIFFERENT GROUPS OF TNPB NUCLEASES AND ARE LIKELY TO BE EVOLUTIONARY INTERMEDIATES ON THE PATH FROM
TNPB TO BONA FIDE CLASS 2 CRISPR EFFECTORS. Article CAS PubMed PubMed Central Google Scholar * Peters, J. E., Makarova, K. S., Shmakov, S. & Koonin, E. V. Recruitment of CRISPR-Cas
systems by Tn7-like transposons. _Proc. Natl Acad. Sci. USA_ 114, E7358–E7366 (2017). THIS ARTICLE IS THE FIRST DESCRIPTION OF DERIVED CRISPR–CAS SYSTEMS CARRIED BY TN7-LIKE TRANSPOSONS. A
HYPOTHETICAL MECHANISM FOR CRRNA-GUIDED TRANSPOSITION IS PROPOSED. Article CAS PubMed Google Scholar * McDonald, N. D., Regmi, A., Morreale, D. P., Borowski, J. D. & Fidelma Boyd, E.
CRISPR-Cas systems are present predominantly on mobile genetic elements in Vibrio species. _BMC Genomics_ 20, 105 (2019). Article PubMed PubMed Central Google Scholar * Ozcan, A. et al.
Type IV CRISPR RNA processing and effector complex formation in Aromatoleum aromaticum. _Nat. Microbiol._ 4, 89–96 (2019). THIS ARTICLE PRESENTS THE MOST THOROUGH AVAILABLE CHARACTERIZATION
OF THE STRUCTURE AND BIOCHEMICAL ACTIVITIES OF TYPE IV CRISPR–CAS SYSTEMS. THE SIMILARITY OF THE EFFECTOR COMPLEX STRUCTURE TO THOSE OF TYPE I IS DEMONSTRATED, SUGGESTING THAT TYPE IV IS AN
EXTREMELY DERIVED FORM OF TYPE I. Article PubMed Google Scholar * Maier, L. K., Dyall-Smith, M. & Marchfelder, A. The adaptive immune system of Haloferax volcanii. _Life (Basel)_ 5,
521–537 (2015). Google Scholar * Seed, K. D., Lazinski, D. W., Calderwood, S. B. & Camilli, A. A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate
immunity. _Nature_ 494, 489–491 (2013). Article CAS PubMed PubMed Central Google Scholar * Naser, I. B. et al. Analysis of the CRISPR-Cas system in bacteriophages active on epidemic
strains of _Vibrio cholerae_ in Bangladesh. _Sci. Rep._ 7, 14880 (2017). Article PubMed PubMed Central Google Scholar * Roberts, A. P. & Mullany, P. Tn916-like genetic elements: a
diverse group of modular mobile elements conferring antibiotic resistance. _FEMS Microbiol. Rev._ 35, 856–871 (2011). Article CAS PubMed Google Scholar * Parks, A. R. et al.
Transposition into replicating DNA occurs through interaction with the processivity factor. _Cell_ 138, 685–695 (2009). Article CAS PubMed PubMed Central Google Scholar * Harrington, L.
B. et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. _Science_ 362, 839–842 (2018). THIS ARTICLE IS AN EXPERIMENTAL VALIDATION OF THE INTERFERENCE ACTIVITY OF THREE SMALL
TYPE V EFFECTOR PROTEINS THAT ARE CLOSELY RELATED TO TNPB AND SOME OF THE SUBTYPE V-U VARIANTS DESCRIBED IN SHMAKOV ET AL. ( _NAT. REV. MICROBIOL._ , 2017). PREFERENTIAL ACTIVITY AGAINST
SINGLE-STRANDED DNA, AS OPPOSED TO DOUBLE-STRANDED DNA, AS IS THE CASE FOR CAS12, IS DEMONSTRATED. THE CORRESPONDING CRISPR–CAS TYPE SYSTEMS ARE NOW CLASSIFIED AS SUBTYPE V-F. Article CAS
PubMed Google Scholar * Yan, W. X. et al. Functionally diverse type V CRISPR-Cas systems. _Science_ 363, 88–91 (2019). THIS WORK COMPLEMENTS HARRINGTON ET AL. (2018) BY DEMONSTRATING THE
ACTIVITY OF A DISTINCT V-U VARIANT (RECLASSIFIED SUBTYPE V-G) THAT UNEXPECTEDLY SHOWS STRONG PREFERENCE FOR SINGLE-STRANDED RNA SUBSTRATES. Article CAS PubMed Google Scholar * He, S. et
al. The IS200/IS605 family and “peel and paste” single-strand transposition mechanism. _Microbiol. Spectr_. https://doi.org/10.1128/microbiolspec.MDNA3-0039-2014 (2015). Article PubMed
Google Scholar * Chen, J. S. et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. _Science_ 360, 436–439 (2018). Article CAS PubMed Google Scholar
* Choi, K. Y., Spencer, J. M. & Craig, N. L. The Tn7 transposition regulator TnsC interacts with the transposase subunit TnsB and target selector TnsD. _Proc. Natl Acad. Sci. USA_ 111,
E2858–E2865 (2014). Article CAS PubMed Google Scholar * Peters, J. E. Tn7. _Microbiol. Spectr_. https://doi.org/10.1128/microbiolspec.MDNA3-0010-2014 (2014). Article PubMed Google
Scholar * Koonin, E. V. & Krupovic, M. Evolution of adaptive immunity from transposable elements combined with innate immune systems. _Nat. Rev. Genet._ 16, 184–192 (2015). Article CAS
PubMed Google Scholar * Nowacki, M., Shetty, K. & Landweber, L. F. RNA-mediated epigenetic programming of genome rearrangements. _Annu. Rev. Genomics Hum. Genet._ 12, 367–389 (2011).
Article CAS PubMed PubMed Central Google Scholar * Newire, E., Aydin, A., Juma, S., Enne, V. & Roberts, A. P. Identification of a type IV CRISPR-Cas system located exclusively on
IncHI1B/ IncFIB plasmids in Enterobacteriaceae. Preprint at _bioRxiv_ https://doi.org/10.1101/536375 (2019) Article Google Scholar * Carroll, K. S. et al. A conserved mechanism for
sulfonucleotide reduction. _PLOS Biol._ 3, e250 (2005). Article PubMed PubMed Central Google Scholar * You, D., Wang, L., Yao, F., Zhou, X. & Deng, Z. A novel DNA modification by
sulfur: DndA is a NifS-like cysteine desulfurase capable of assembling DndC as an iron-sulfur cluster protein in _Streptomyces lividans_. _Biochemistry_ 46, 6126–6133 (2007). Article CAS
PubMed Google Scholar * Makarova, K. S., Wolf, Y. I. & Koonin, E. V. Comparative genomics of defense systems in archaea and bacteria. _Nucleic Acids Res._ 41, 4360–4377 (2013). Article
CAS PubMed PubMed Central Google Scholar * Simon, N. C., Aktories, K. & Barbieri, J. T. Novel bacterial ADP-ribosylating toxins: structure and function. _Nat. Rev. Microbiol._ 12,
599–611 (2014). Article CAS PubMed PubMed Central Google Scholar * Labrie, S. J., Samson, J. E. & Moineau, S. Bacteriophage resistance mechanisms. _Nat. Rev. Microbiol._ 8, 317–327
(2010). Article CAS PubMed Google Scholar * Shabbir, M. A. et al. Bacteria versus bacteriophages: parallel evolution of immune arsenals. _Front. Microbiol._ 7, 1292 (2016). Article
PubMed PubMed Central Google Scholar * Villion, M. & Moineau, S. The double-edged sword of CRISPR-Cas systems. _Cell Res._ 23, 15–17 (2013). Article CAS PubMed Google Scholar *
Angermeyer, A., Das, M. M., Singh, D. V. & Seed, K. D. Analysis of 19 highly conserved _Vibrio cholerae_ bacteriophages isolated from environmental and patient sources over a twelve-year
period. _Viruses_ 10, E299 (2018). Article PubMed Google Scholar * Al-Shayeb, B. et al. Clades of huge phage from across Earth’s ecosystems. Preprint at _bioRxiv_
https://doi.org/10.1101/572362 (2019). Article Google Scholar * Hooton, S. P., Brathwaite, K. J. & Connerton, I. F. The bacteriophage carrier state of _Campylobacter jejuni_ features
changes in host non-coding RNAs and the acquisition of new host-derived CRISPR spacer sequences. _Front. Microbiol._ 7, 355 (2016). Article PubMed PubMed Central Google Scholar * Hooton,
S. P. & Connerton, I. F. _Campylobacter jejuni_ acquire new host-derived CRISPR spacers when in association with bacteriophages harboring a CRISPR-like Cas4 protein. _Front. Microbiol._
5, 744 (2014). PubMed Google Scholar * He, F. et al. Anti-CRISPR proteins encoded by archaeal lytic viruses inhibit subtype I-D immunity. _Nat. Microbiol._ 3, 461–469 (2018). Article CAS
PubMed Google Scholar * Koonin, E. V. & Makarova, K. S. Anti-CRISPRs on the march. _Science_ 362, 156–157 (2018). Article CAS PubMed Google Scholar * Sebaihia, M. et al. The
multidrug-resistant human pathogen _Clostridium difficile_ has a highly mobile, mosaic genome. _Nat. Genet._ 38, 779–786 (2006). Article PubMed Google Scholar * Minot, S. et al. The human
gut virome: inter-individual variation and dynamic response to diet. _Genome Res._ 21, 1616–1625 (2011). Article CAS PubMed PubMed Central Google Scholar * Garcia-Heredia, I. et al.
Reconstructing viral genomes from the environment using fosmid clones: the case of haloviruses. _PLOS ONE_ 7, e33802 (2012). Article CAS PubMed PubMed Central Google Scholar * Faure, G.
et al. Comparative genomics and evolution of _trans_-activating RNAs in class 2 CRISPR-Cas systems. _RNA Biol_. https://doi.org/10.1080/15476286.2018.1493331 (2018). Article PubMed PubMed
Central Google Scholar * Shmakov, S. A. et al. The CRISPR spacer space is dominated by sequences from species-specific mobilomes. _mBio_ 8, e01397-17 (2017). Article CAS PubMed PubMed
Central Google Scholar * Anderson, E. M. et al. Systematic analysis of CRISPR-Cas9 mismatch tolerance reveals low levels of off-target activity. _J. Biotechnol._ 211, 56–65 (2015). Article
CAS PubMed Google Scholar * Zheng, T. et al. Profiling single-guide RNA specificity reveals a mismatch sensitive core sequence. _Sci. Rep._ 7, 40638 (2017). Article CAS PubMed PubMed
Central Google Scholar * Kleinstiver, B. P. et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. _Nature_ 523, 481–485 (2015). Article PubMed PubMed Central Google
Scholar * Horvath, P. et al. Diversity, activity, and evolution of CRISPR loci in _Streptococcus thermophilus_. _J. Bacteriol._ 190, 1401–1412 (2008). Article CAS PubMed Google Scholar
* Leenay, R. T. et al. Identifying and visualizing functional PAM diversity across CRISPR-Cas systems. _Mol. Cell_ 62, 137–147 (2016). Article CAS PubMed PubMed Central Google Scholar *
Zhang, Y. et al. Processing-independent CRISPR RNAs limit natural transformation in _Neisseria meningitidis_. _Mol. Cell_ 50, 488–503 (2013). Article CAS PubMed PubMed Central Google
Scholar * Amitai, G. & Sorek, R. CRISPR-Cas adaptation: insights into the mechanism of action. _Nat. Rev. Microbiol._ 14, 67–76 (2016). Article CAS PubMed Google Scholar * Pawluk,
A. et al. Naturally occurring off-switches for CRISPR-Cas9. _Cell_ 167, 1829–1838 (2016). Article CAS PubMed PubMed Central Google Scholar * Pawluk, A., Davidson, A. R. & Maxwell,
K. L. Anti-CRISPR: discovery, mechanism and function. _Nat. Rev. Microbiol._ 16, 12–17 (2018). Article CAS PubMed Google Scholar * Maxwell, K. L. The anti-CRISPR story: a battle for
survival. _Mol. Cell_ 68, 8–14 (2017). Article CAS PubMed Google Scholar * Varble, A., Meaden, S., Barrangou, R., Westra, E. R. & Marraffini, L. A. Recombination between phages and
CRISPR-cas loci facilitates horizontal gene transfer in staphylococci. _Nat. Microbiol._ https://doi.org/10.1038/s41564-019-0400-2 (2019). Article PubMed Google Scholar * Koonin, E. V.
& Krupovic, M. A movable defense. _TheScientist_ https://www.the-scientist.com/features/a-movable-defense-36135 (2015). * Arndt, D. et al. PHASTER: a better, faster version of the PHAST
phage search tool. _Nucleic Acids Res._ 44, W16–W21 (2016). Article CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS G.F., S.A.S., K.S.M. and E.V.K. are
supported by funds from the Intramural Research Program of the National Institutes of Health of the USA. S.A.S. was additionally supported by the Russian Foundation for Basic Research
(research project 18-34-00012) and a systems biology fellowship from Philip Morris Sales and Marketing. J.E.P. was supported by the US Department of Agriculture National Institute of Food
and Agriculture Hatch Project NYC-189438. D.R.C., W.X.Y. and D.A.S. are supported by Arbor Biotechnologies. REVIEWER INFORMATION _Nature Reviews Microbiology_ thanks U. Gophna, and other
anonymous reviewer(s), for their contribution to the peer review of this work. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * National Center for Biotechnology Information, National Library
of Medicine, National Institutes of Health, Bethesda, MD, USA Guilhem Faure, Sergey A. Shmakov, Kira S. Makarova & Eugene V. Koonin * Broad Institute of MIT and Harvard, Cambridge, MA,
USA Guilhem Faure * Skolkovo Institute of Science and Technology, Skolkovo, Russia Sergey A. Shmakov * Arbor Biotechnologies, Cambridge, MA, USA Winston X. Yan, David R. Cheng & David A.
Scott * Department of Microbiology, Cornell University, Ithaca, NY, USA Joseph E. Peters Authors * Guilhem Faure View author publications You can also search for this author inPubMed Google
Scholar * Sergey A. Shmakov View author publications You can also search for this author inPubMed Google Scholar * Winston X. Yan View author publications You can also search for this
author inPubMed Google Scholar * David R. Cheng View author publications You can also search for this author inPubMed Google Scholar * David A. Scott View author publications You can also
search for this author inPubMed Google Scholar * Joseph E. Peters View author publications You can also search for this author inPubMed Google Scholar * Kira S. Makarova View author
publications You can also search for this author inPubMed Google Scholar * Eugene V. Koonin View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS
G.F., S.A.S., W.X.Y., D.R.C., D.A.S., J.E.P., K.S.M. and E.V.K. researched the data for the article. G.F., W.X.Y., D.R.C., D.A.S., J.E.P., K.S.M. and E.V.K. substantially contributed to the
discussion of the content. E.V.K. wrote the article. G.F., W.X.Y., D.R.C., D.A.S., J.E.P., K.S.M. and E.V.K. reviewed and edited the manuscript before submission. CORRESPONDING AUTHOR
Correspondence to Eugene V. Koonin. ETHICS DECLARATIONS COMPETING INTERESTS D.R.C., W.X.Y. and D.A.S. are shareholders of Arbor Biotechnologies. All other authors declare no competing
interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY
INFORMATION SUPPLEMENTARY INFORMATION SUPPLEMENTARY DATASET 1 SUPPLEMENTARY DATASET 2 SUPPLEMENTARY DATASET 3 SUPPLEMENTARY DATASET 4 SUPPLEMENTARY DATASET 5 GLOSSARY * CRISPR spacers Unique
sequences of 20–60 nucleotides inserted between the repeats in the CRISPR array and employed, as part of the CRISPR RNA, for targeting DNA molecules containing a homologous protospacer. *
CRISPR array A series of direct repeats in bacterial and archaeal genomes interspersed with spacers that are acquired primarily from mobile genetic element DNA. * crRNAs Short RNAs, produced
by processing of the primary transcript of a CRISPR array, that consists of a spacer and portions of the flanking repeats and functions as a guide to target DNA or RNA molecules containing
cognate protospacers. * Mini-arrays Minimal forms of a CRISPR array that consists of a proximal repeat, a spacer and a distal repeat, or, more commonly, a partial repeat; so far identified
in virus and provirus genomes. * Solitary repeat units (SRUs). Short sequences, so far identified in virus and provirus genomes, that are (nearly) identical to a repeat from a CRISPR array.
* CRISPR adaptation module A group of _cas_ genes dedicated to the selection and insertion of new spacers into CRISPR arrays. * TnpB A nuclease containing a RuvC-like domain that is encoded
by numerous transposons (insertion sequences) although not required for transposition. * IS605-like transposons A family of bacterial and archaeal insertion sequence elements that encode a
distinct transposase (TnpA) and often a second nuclease (TnpB) that is also found in numerous non-autonomous IS605-like transposons lacking TnpA. * Tn7-like transposons A derivative of Tn7
family transposons lacking some accessory genes involved in transposition and in some cases carrying derived CRISPR–Cas systems lacking the interference capacity. * CRISPR effector module A
suite of Cas proteins (Class 1 CRISPR–Cas systems) or a single large protein (Class 2 CRISPR–Cas systems) that are responsible for maturation of the CRISPR RNA and interference. * R-loops
Three-stranded structures that consist of a DNA–RNA hybrid and the displaced single-stranded DNA, formed during transcription and other processes including target recognition by CRISPR–Cas
effector complexes (proteins). * Protospacer A piece of DNA, typically from a mobile genetic element genome, that is inserted into a CRISPR array by the CRISPR adaptation complex, to become
a spacer. * CysH enzymes Enzymes of the adenosine 5′-phosphosulfate reductase family that reduce activated sulfate to sulfite; associated with many type IV CRISPR–Cas systems. * Exaptation
Co-option (recruitment) of a biological entity, such as a protein or DNA sequence, for a role that is distinct from its original function. * Anti-CRISPR proteins Diverse proteins encoded by
many bacterial and archaeal viruses that inhibit the host CRISPR–Cas systems, typically by binding and inactivating the effector complex (protein). * _Trans_activating RNAs (tracrRNAs). RNA
molecules encoded by all known type II CRISPR–Cas systems and some type V systems that consist of a sequence partially complementary to the corresponding repeat and a unique portion;
co-folding of the tracrRNA with the pre-crRNA is essential for crRNA maturation and interference by the respective CRISPR–Cas systems. * Protospacer adjacent motif (PAM). A short,
two-to-three-nucleotide motif, the presence of which next to the protospacer sequence is essential for both adaptation and interference by most of the CRISPR–Cas systems; different PAM
sequences are required by different CRISPR-Cas types and subtypes. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Faure, G., Shmakov, S.A., Yan, W.X.
_et al._ CRISPR–Cas in mobile genetic elements: counter-defence and beyond. _Nat Rev Microbiol_ 17, 513–525 (2019). https://doi.org/10.1038/s41579-019-0204-7 Download citation * Published:
05 June 2019 * Issue Date: August 2019 * DOI: https://doi.org/10.1038/s41579-019-0204-7 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get
shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative