Play all audios:
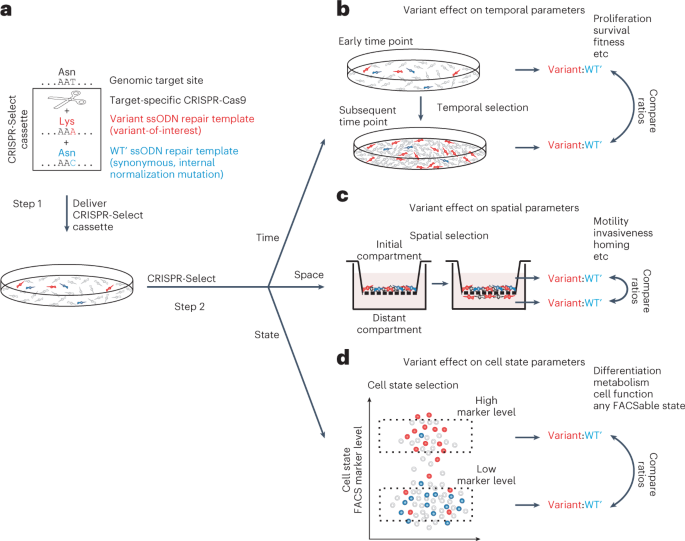
We developed a CRISPR-based functional assay for genetic sequence variants found in human disease, probing their effects on cell proliferation, survival, motility and any physiological or
pathological process measurable by fluorescence-activated cell sorting (FACS). The assay accurately assessed variant pathogenicity, drug responsiveness or resistance and mechanistic role in
disease, in vitro and in vivo. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your
institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $29.99 / 30 days cancel any time Learn more Subscribe to this journal
Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices
may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support
REFERENCES * Landrum, M. J. et al. ClinVar: Improving access to variant interpretations and supporting evidence. _Nucleic Acids Res._ 48, pD835–D844 (2020). AN ARTICLE ON CLINVAR, AN
EXHAUSTIVE DATABASE FOR HUMAN GENETIC VARIANTS AND INTERPRETATIONS OF THEIR ROLE IN DISEASE. Article Google Scholar * Richards, S. et al. Standards and guidelines for the interpretation of
sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. _Genet. Med._ 17, 405–424 (2015).
GUIDELINES OF THE CLINICAL GENETICS COMMUNITY FOR ASSESSING THE PATHOGENICITY OF GENETIC SEQUENCE VARIANTS. Article Google Scholar * Burke, W. et al. The challenge of genetic variants of
uncertain clinical significance. _Ann. Intern. Med._ 175, 994–1000 (2022). A REVIEW DISCUSSING THE PROBLEMS OF VUS FINDINGS FOR GENETIC TESTING LABORATORIES, CLINICIANS AND PATIENTS. Article
Google Scholar * Brnich, S. E. et al. Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant framework. _Genome Med_ 12, 3
(2019). AN ARTICLE THAT ASSESSES FUNCTIONAL ASSAYS FOR VARIANT INTERPRETATION, PROVIDING RECOMMENDATIONS AND GUIDELINES FOR THEIR USE. Article Google Scholar * Pickar-Oliver, A. &
Gersbach, C. A. The next generation of CRISP–Cas technologies and applications. _Nat. Rev. Mol. Cell. Biol._ 20, 490–507 (2019). A REVIEW ARTICLE ON THE MECHANISMS AND APPLICATIONS OF
CRISPR–CAS GENOME-EDITING TECHNOLOGY. Article CAS Google Scholar Download references ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional
claims in published maps and institutional affiliations. THIS IS A SUMMARY OF: Niu, Y. et al. Multiparametric and accurate functional analysis of genetic sequence variants using
CRISPR-Select. _Nat. Genet_. https://doi.org/10.1038/s41588-022-01224-7 (2022). RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE A versatile functional
assay for genetic variants in human disease. _Nat Genet_ 54, 1778–1779 (2022). https://doi.org/10.1038/s41588-022-01226-5 Download citation * Published: 05 December 2022 * Issue Date:
December 2022 * DOI: https://doi.org/10.1038/s41588-022-01226-5 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a
shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative