Play all audios:
ABSTRACT This dataset comprises 38 breast ultrasound scans from patients, encompassing a total of 683 images. The scans were conducted using a Siemens ACUSON S2000TM Ultrasound System from
2022 to 2023. The dataset is specifically created for the purpose of segmenting breast lesions, with the goal of identifying the area and contour of the lesion, as well as classifying it as
either benign or malignant. The images can be classified into three categories based on their findings: 419 are normal, 174 are benign, and 90 are malignant. The ground truth is given as RGB
segmentation masks in individual files, with black indicating normal breast tissue and green and red indicating benign and malignant lesions, respectively. This dataset enables researchers
to construct and evaluate machine learning models for identifying between benign and malignant tumours in authentic breast ultrasound images. The segmentation annotations provided by expert
radiologists enable accurate model training and evaluation, making this dataset a valuable asset in the field of computer vision and public health. SIMILAR CONTENT BEING VIEWED BY OTHERS
CURATED BENCHMARK DATASET FOR ULTRASOUND BASED BREAST LESION ANALYSIS Article Open access 31 January 2024 SEMIAUTOMATED SEGMENTATION OF BREAST TUMOR ON AUTOMATIC BREAST ULTRASOUND IMAGE
USING A LARGE-SCALE MODEL WITH CUSTOMIZED MODULES Article Open access 19 May 2025 SEGMENT ANYTHING IN MEDICAL IMAGES Article Open access 22 January 2024 BACKGROUND & SUMMARY While there
are quite a few computer-aided detection (CAD) systems for mammography used in radiology for breast cancer screening, the same cannot be said for CAD systems using breast ultrasound images
(BUS). Mammography CAD systems use a range of techniques, including conventional machine learning approaches1 and deep learning methods2. Implementing a screening system that incorporates
diagnostic tools readily available in most clinical centres, like ultrasound imaging devices, would be a significant breakthrough in combating breast cancer. This system has the potential to
greatly improve breast cancer prognosis and reduce breast mortality due to cancer. Precise detection and segmentation of abnormalities in breast ultrasound images, including both benign and
malignant tumours, is essential for the diagnosis of suspicious masses detected in mammograms and the assessment of dense breasts3. Although there are many computer-aided detection (CAD)
systems for mammography used in radiology for breast cancer screening, CAD systems for breast ultrasound images (BUS) are relatively rare. An exception is the S-Detect™ system offered on
Samsung ultrasound machines. This software analyzes breast lesions and classifies them according to the BI-RADS® ATLAS. This system can improve the accuracy and reliability of breast cancer
detection through ultrasound. Nevertheless, despite the advancements made, numerous challenges must be tackled in order to develop robust machine learning models for breast ultrasound
analysis. One major challenge is the availability of large and well-annotated datasets. High-quality datasets are crucial for the training and validation of machine learning models. In order
to ensure that the models can be applied effectively to real-world situations, that is, to ensure they generalize well to real-world scenarios, it is necessary for them to include a wide
range of examples of breast lesions, encompassing different types and stages of cancer. The dataset introduced herein (BUS-UCLM dataset) aims to provide a comprehensive resource for the
development and evaluation of machine learning algorithms for breast ultrasound image analysis. This dataset comprises a wide range of breast ultrasound images, alongside detailed
annotations provided by expert radiologists. The dataset is designed to support research in a range of tasks, including lesion segmentation, classification, and detection. METHODS IMAGE
ACQUISITION AND ANONYMIZATION Ultrasound images were collected from 2022 to 2023 at Ciudad Real General University Hospital. Images were acquired using the Siemens Acuson S2000 ultrasound
system, with the 18L6 HD probe, and using the standard beamforming method. The pixel resolution varied across the dataset, with the most common resolution being (0.0639205, 0.0639205). Other
resolutions were also present but for a smaller number of images, including (0.0568182, 0.0568182) for 31 images, (0.0710227, 0.0710227) for 51 images, and several other resolutions each
represented by fewer than 20 images. The dimensions of each image were 768 × 1024 pixels. The dataset is derived from authentic clinical studies, without any predefined selection criteria
for patients or images. This approach ensures a realistic representation of clinical scenarios. The study was reviewed and approved by the Ethics Committee of Ciudad Real General University
Hospital as part of project PID2021-127567NB-I00. In addition, informed consent was obtained from all participants to collect and share the data. Participants were assured that their
confidentiality would be maintained. Images were initially stored in DICOM format and subsequently converted to PNG files. To ensure patient privacy and comply with data protection
regulations, folders and DICOM files were renamed using random four-letter sequences, with each sequence uniquely identifying images from the same patient. This approach enables
subject-based cross-validation partitions. Additionally, sensitive data in the DICOM header fields was anonymized, and a YOLOv8-trained CNN was employed for text detection to mask sensitive
information in the DICOM pixel data by overlaying black rectangles4. IMAGE ANNOTATION A diverse dataset comprising both benign and malignant cases was compiled. The malignancy status of each
lesion was verified through biopsy procedures. Expert radiologists then meticulously produced manual annotations. The dataset was labeled with segmentation masks, which made it suitable for
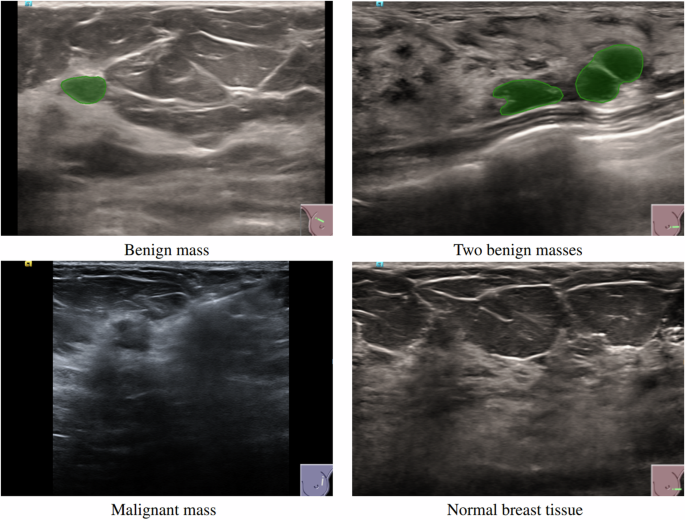
semantic segmentation, instance segmentation or detection tasks. It was noted that in other publicly available datasets, only one lesion per image was labeled, even if more than one lesion
was present. Since having scans with more than one lesion is common, especially when dealing with breast cysts—which represent 25% of all breast masses and often appear in clusters—all
lesions appearing in an image were labeled. One radiologist delineated the lesion contours, which, coupled with diagnostic information, facilitated the creation of precise segmentation
masks. Additionally, another radiologist reviewed and approved the marks, both reaching a consensus on the delineation. For the annotation generation, we employed CVAT, an Open Data
Annotation Platform, to manually generate the ground truth annotations5. All processed images are available in PNG format. Furthermore, scans of normal tissue were included. As it is common
for most of the breast tissue to be normal in an ultrasound session, these samples are important to measure the number of false positives, particularly in the test phase. Some samples with
the segmentation masks overlaid are shown in Fig. 1. Multiple images were collected for each patient, taken from different breast cross-sections to ensure comprehensive coverage of the area
of interest. Therefore, these images are 2D cross-sectional views (Fig. 2). DATA PROTECTION AND COMPLIANCE To ensure compliance with legal and regulatory frameworks for medical data, we
implemented data protection measures in accordance with the General Data Protection Regulation (GDPR) and the Health Insurance Portability and Accountability Act (HIPAA). Specifically, we
adhered to both GDPR Article 9 and HIPAA’s Privacy Rule, which mandate the protection of personal data, by ensuring that all identifiable information was removed from the dataset. This
included both direct identifiers such as names and dates, and indirect identifiers such as unique codes that could be traced back to an individual. These measures ensured that the data could
not be used to identify individuals, either on its own or in combination with other information. These safeguards were applied to the initial DICOM files, ensuring that even the authors do
not retain sensitive data. All anonymized data was handled and stored securely to prevent unauthorized access. Furthermore, only PNG files are publicly shared, which strengthens compliance
with data privacy regulations. The anonymization script, along with other auxiliary code, has been uploaded to a GitHub repository to ensure transparency and reproducibility of our methods
(https://github.com/noeliavallez/BUS-UCLM-Dataset). DATA RECORDS The dataset is available at Mendeley Data6. It is organized into a main folder that includes two subfolders and a CSV file
with image information, including the image name, resolution, label, and whether it was acquired with Doppler, is a combined image, or has masks. One subfolder holds the ultrasound images in
PNG format and the other one the segmentation masks. The images are named using a pattern of four random letters followed by an underscore and a sequence number (e.g., XXXX_YYY.png), where
’XXXX’ represents an anonymized patient identifier and ’YYY’ is the image number within that patient’s study. The segmentation masks utilize a color-coding system in RGB format to indicate
different types of tissue: red (255,0,0) for malignant lesions, green (0,255,0) for benign lesions, and black (0,0,0) for normal breast tissue and other non-lesion areas. A visual
representation of the dataset’s structure is provided in Fig. 3, offering an overview of how the data is organized and labeled. Table 1 contains the number of images per patient. The dataset
is publicly available and free to use for research purposes, fostering collaboration and innovation in the field of breast ultrasound analysis. The dataset is licensed under CC-BY 4.0.
TECHNICAL VALIDATION The dataset was compiled from ultrasound images acquired at the Ciudad Real General University Hospital between 2022 and 2023. Each image underwent a conversion from
DICOM to PNG format, a process meticulously designed to strip away any identifiable patient information, thereby upholding the strictest standards of privacy and confidentiality. Two expert
radiologists with extensive experience in breast imaging annotated the dataset. Both radiologists reached a consensus on the annotations. This consensus is crucial because it ensures that
the annotations are accurate and reliable, reducing the likelihood of individual bias or error. The malignancy of the lesions was confirmed through biopsy. This direct correlation between
the biopsy results and the annotations ensures that the dataset reflects the highest level of precision and accuracy for lesion classification. The BUS-UCLM includes 683 images collected
from 38 patients, comprising 419 normal, 174 benign, and 90 malignant images. A comparison with other existing public datasets has been made. Generally, the BUS-UCLM dataset is similar to
these datasets but distinguishes itself by including samples without lesions and samples with multiple lesions labelled. A summary of relevant features is presented in Table 3. * The Breast
Ultrasound Images (BUSI) dataset contains 780 images annotated for segmentation, including 437 benign, 210 malignant, and 133 normal images. It is a widely used dataset but has limitations
in the variety of images and the overall size of the dataset, which may not be sufficient for training highly generalizable models. * The UDIAT DatasetB, used in several studies, contains
163 annotated images with 109 benign and 54 malignant cases. This dataset is relatively small and does not include normal tissue images. Furthermore, it is not publicly downloadable, which
limits its accessibility for broader research use, although it can be obtained by contacting the authors. * The RODTOOK dataset includes 144 images with 57 benign and 87 malignant lesions.
Although it is useful for segmentation tasks, its smaller size and lack of normal tissue samples present limitations for comprehensive model training and evaluation. * The Open Access Series
of Breast Ultrasound Data (OASBUD) provides 200 images with an equal split between benign and malignant cases. This dataset is publicly available and has been used in numerous studies.
However, as with other datasets, it does not include normal tissue scans. The image quality of BUS-UCLM is superior to that of other datasets since it was gathered with recent equipment,
more robust to the image noise inherent in ultrasound imaging. Figure 4 shows examples of the 5 datasets with their corresponding segmentation masks. To validate the dataset, a UNet model
was trained using the BUS-UCLM dataset to segment image pixels into background, benign, and malignant categories. Ninety percent of the cases were used for training, and ten percent for
testing, with partitions done at the patient level. The model achieved a Dice score of 0.68, indicating that despite the small size, the dataset is sufficient to develop models with
reasonable performance (Fig. 5). This highlights the great potential of this dataset. When combined with other publicly available BUS segmentation datasets, it may significantly enhance the
reliability and predictive performance of the model across diverse cases. Integrating our dataset with those listed in Table 3 may be particularly beneficial. As a result, despite the modest
sample size, the rich quality of the data and the careful segmentation procedures provide a solid foundation for training and evaluating machine learning models. This approach ensures that
the findings are relevant and meaningful, with the potential for scalability once additional datasets are integrated. Further work will focus on expanding the dataset to include more
examples, which will further improve the model’s performance. IDENTIFICATION OF BIASES The potential sources of bias of the use of the BUS-UCLM dataset have been identified. One of them is
the demographic bias. The dataset was collected exclusively from the General University Hospital of Ciudad Real in Spain. This localized collection process may introduce demographic bias, as
the patient population may not represent broader geographical or ethnic groups. This limitation could impact the applicability of the model to populations with different genetic, ethnic, or
lifestyle factors. Clinical bias is another concern. The dataset primarily includes cases verified through biopsy and annotated by two expert radiologists. While this ensures high-quality
annotations, it may limit variability in the dataset, as diagnoses made by general practitioners or less experienced clinicians are not represented. This could lead to a model that is overly
dependent on expert-level data, reducing its robustness in real-world scenarios where data quality and diagnostic expertise vary. Selection bias may also arise as a result of the limited
dataset size. Despite not having a predefined inclusion criteria, the dataset includes only 38 patients. This small sample size may result in underrepresentation of certain lesion types or
clinical conditions, reducing the diversity of the dataset. Finally, some images in the dataset contain annotations such as arrows, yellow crosses, and bounding boxes that overlap with the
mass areas. These marks may influence the outcome of the models, introducing bias. Additionally, some images are Doppler ultrasound images, and others are combined images containing multiple
scans in one. To mitigate these problems, we have provided a CSV file with detailed metadata for each image, including three columns indicating whether the image contains Doppler features,
visual marks, or is a combined image. This allows users to include or exclude specific subsets of images based on their needs. COMBINATION WITH OTHER DATASETS To assess the impact of
combining the BUS-UCLM dataset with other datasets, we integrated all datasets listed in Table 3 and trained five segmentation models (UNet, AttUnet, SK-UNet, DeepLabv3, and Mask R-CNN) with
the five datasets compared in this work, extending the work of Cory _et al_. from binary segmentation to three classes (background, benign, and malignant)7. A 5-fold cross-validation was
employed, ensuring that the folds were the same for all models and that partitions were made at patient level and not at image level to prevent having images from the same patient in more
than one fold. Intersection over union (IoU), accuracy (Acc), Dice score, Precision, and Recall were calculated. The results presented in Table 2 are the averaged results of all folds. From
the five models tested, Mask R-CNN outperformed other networks with the highest average scores across all metrics: IoU=65.46%, Acc=74.38%, Dice=77.09%, Precision=80.29%, and Recall=74.38%.
These results are in line with those presented in the literature7. These results suggest that the dataset does not introduce significant biases that could negatively affect model
performance. However, to enhance the generalizability of diagnostic algorithms, we recommend using this dataset in conjunction with other resources. Although other available datasets are
also small and may share similar biases, integrating multiple datasets can still help create more comprehensive models. This approach captures a broader spectrum of real-world features,
ultimately improving robustness and applicability. CODE AVAILABILITY A source code repository is available on GitHub (https://github.com/noeliavallez/BUS-UCLM-Dataset) with useful scripts to
convert the masks to the COCO annotation format in JSON and overlay the segmented areas onto the original images. REFERENCES * Vallez, N., Bueno, G., Deniz, O., Dorado, J., Seoane, J. A.,
Pazos, A. & Pastor, C. Breast density classification to reduce false positives in CADe systems. _Computer methods and programs in biomedicine_ 113, 569–584 (2014). Article PubMed
Google Scholar * Zhong, Y., Piao, Y., Tan, B. & Liu, J. A multi-task fusion model based on a residual–multi-layer perceptron network for mammographic breast cancer screening. Computer
Methods and Programs in Biomedicine108101 (2024). * Kolb, T. M., Lichy, J. & Newhouse, J. H. Comparison of the performance of screening mammography, physical examination, and breast US
and evaluation of factors that influence them: an analysis of 27,825 patient evaluations. _Radiology_ 225, 165–175 (2002). Article PubMed Google Scholar * Singh, A. _et al_. TextOCR:
Towards large-scale end-to-end reasoning for arbitrary-shaped scene text (2021). * Roboflow. CVAT: Open Data Annotation Platform. https://app.cvat.ai. Accessed: 2024-06-17. * Vallez, N.,
Bueno, G., Deniz, O., Rienda, M. A. & Pastor, C. BUS-UCLM: Breast ultrasound lesion segmentation dataset. Mendeley Data V3, https://doi.org/10.17632/7fvgj4jsp7.3 (2025). * Thomas, C.,
Byra, M., Marti, R., Yap, M. H. & Zwiggelaar, R. BUS-Set: A benchmark for quantitative evaluation of breast ultrasound segmentation networks with public datasets. _Medical Physics_ 50,
3223–3243, https://doi.org/10.1002/mp.16287 (2023). Article ADS CAS PubMed MATH Google Scholar * Al-Dhabyani, W., Gomaa, M., Khaled, H. & Fahmy, A. Dataset of breast ultrasound
images. _Data in brief_ 28, 104863 (2020). Article PubMed MATH Google Scholar * Yap, Y. S. _et al_. Automated breast ultrasound lesions detection using convolutional neural networks.
_IEEE journal of biomedical and health informatics_ 22, 1218–1226 (2017). Article PubMed Google Scholar * Rodtook, A., Kirimasthong, K., Lohitvisate, W. & Makhanov, S. S. Automatic
initialization of active contours and level set method in ultrasound images of breast abnormalities. _Pattern Recognition_ 79, 172–182 (2018). Article ADS MATH Google Scholar *
Piotrzkowska-Wróblewska, H., Dobruch-Sobczak, K., Byra, M. & Nowicki, A. Open access database of raw ultrasonic signals acquired from malignant and benign breast lesions. _Medical
physics_ 44, 6105–6109 (2017). Article ADS PubMed Google Scholar Download references ACKNOWLEDGEMENTS This work has been funded by the HANS project (Ref. PID2021-127567NB-I00) supported
by the Spanish Ministry of Science, Innovation, and Universities, and by the European Union NextGenerationEU/PRTR. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * VISILAB, E.T.S. Ingeniería
Industrial, University of Castilla-La Mancha, Avda. Camilo José Cela s/n, 13005, Ciudad Real, Spain Noelia Vallez, Gloria Bueno & Oscar Deniz * Hospital General Universitario de Ciudad
Real, C/ Obispo Rafael Torija s/n, 13005, Ciudad Real, Spain Miguel Angel Rienda & Carlos Pastor Authors * Noelia Vallez View author publications You can also search for this author
inPubMed Google Scholar * Gloria Bueno View author publications You can also search for this author inPubMed Google Scholar * Oscar Deniz View author publications You can also search for
this author inPubMed Google Scholar * Miguel Angel Rienda View author publications You can also search for this author inPubMed Google Scholar * Carlos Pastor View author publications You
can also search for this author inPubMed Google Scholar CONTRIBUTIONS Noelia Vallez contributed through conceptualization, data curation, software development, and manuscript writing. Gloria
Bueno and Oscar Deniz acquired funding, administered and supervised the project, and contributed in manuscript reviewing and editing. Miguel A. Rienda and Carlos Pastor participated in data
acquisition, labelling and curation. CORRESPONDING AUTHOR Correspondence to Noelia Vallez. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL
INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. RIGHTS AND PERMISSIONS OPEN ACCESS This
article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction
in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the
licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article
are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and
your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this
licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Vallez, N., Bueno, G., Deniz, O. _et al._ BUS-UCLM: Breast
ultrasound lesion segmentation dataset. _Sci Data_ 12, 242 (2025). https://doi.org/10.1038/s41597-025-04562-3 Download citation * Received: 21 June 2024 * Accepted: 30 January 2025 *
Published: 11 February 2025 * DOI: https://doi.org/10.1038/s41597-025-04562-3 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable
link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative