Play all audios:
ABSTRACT Membrane proteins are designed to fold and function in a lipid membrane, yet folding experiments within a native membrane environment are challenging to design. Here we show that
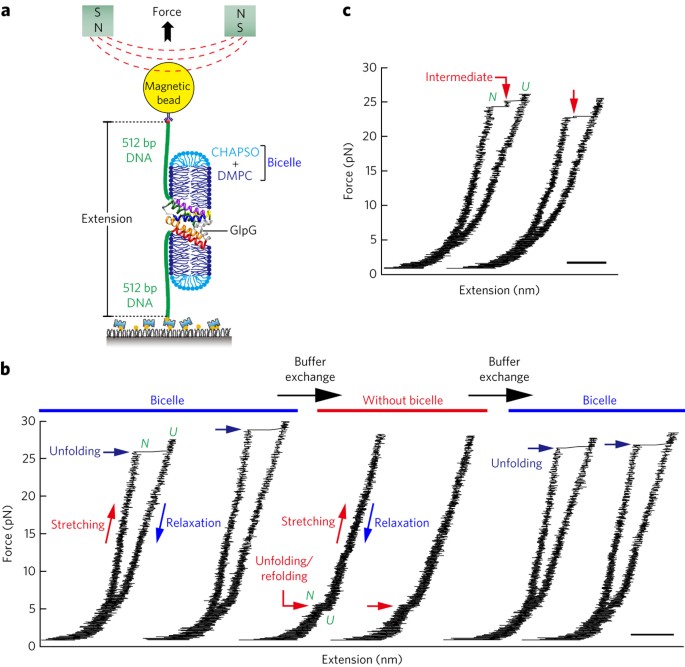
single-molecule forced unfolding experiments can be adapted to study helical membrane protein folding under native-like bicelle conditions. Applying force using magnetic tweezers, we find
that a transmembrane helix protein, _Escherichia coli_ rhomboid protease GlpG, unfolds in a highly cooperative manner, largely unraveling as one physical unit in response to mechanical
tension above 25 pN. Considerable hysteresis is observed, with refolding occurring only at forces below 5 pN. Characterizing the energy landscape reveals only modest thermodynamic stability
(Δ_G_ = 6.5 _k_B_T_) but a large unfolding barrier (21.3 _k_B_T_) that can maintain the protein in a folded state for long periods of time (_t_1/2 ∼3.5 h). The observed energy landscape may
have evolved to limit the existence of troublesome partially unfolded states and impart rigidity to the structure. Access through your institution Buy or subscribe This is a preview of
subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 print issues and online access $259.00 per year only
$21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout
ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS MEMBRANE-MEDIATED PROTEIN
INTERACTIONS DRIVE MEMBRANE PROTEIN ORGANIZATION Article Open access 30 November 2022 FREE ENERGIES OF MEMBRANE STALK FORMATION FROM A LIPIDOMICS PERSPECTIVE Article Open access 15 November
2021 SMALL-RESIDUE PACKING MOTIFS MODULATE THE STRUCTURE AND FUNCTION OF A MINIMAL DE NOVO MEMBRANE PROTEIN Article Open access 16 September 2020 REFERENCES * Engelman, D.M. et al. Membrane
protein folding: beyond the two stage model. _FEBS Lett._ 555, 122–125 (2003). Article CAS PubMed Google Scholar * Bowie, J.U. Solving the membrane protein folding problem. _Nature_ 438,
581–589 (2005). Article CAS PubMed Google Scholar * White, S.H. & von Heijne, G. How translocons select transmembrane helices. _Annu. Rev. Biophys._ 37, 23–42 (2008). Article CAS
PubMed Google Scholar * Hong, H., Blois, T.M., Cao, Z. & Bowie, J.U. Method to measure strong protein-protein interactions in lipid bilayers using a steric trap. _Proc. Natl. Acad.
Sci. USA_ 107, 19802–19807 (2010). Article CAS PubMed PubMed Central Google Scholar * Chang, Y.C. & Bowie, J.U. Measuring membrane protein stability under native conditions. _Proc.
Natl. Acad. Sci. USA_ 111, 219–224 (2014). Article CAS PubMed Google Scholar * Oesterhelt, F. et al. Unfolding pathways of individual bacteriorhodopsins. _Science_ 288, 143–146 (2000).
Article CAS PubMed Google Scholar * Kedrov, A., Janovjak, H., Sapra, K.T. & Muller, D.J. Deciphering molecular interactions of native membrane proteins by single-molecule force
spectroscopy. _Annu. Rev. Biophys. Biomol. Struct._ 36, 233–260 (2007). Article CAS PubMed Google Scholar * Engel, A. & Gaub, H.E. Structure and mechanics of membrane proteins.
_Annu. Rev. Biochem._ 77, 127–148 (2008). Article CAS PubMed Google Scholar * Zocher, M. et al. Single-molecule force spectroscopy from nanodiscs: an assay to quantify folding,
stability, and interactions of native membrane proteins. _ACS Nano_ 6, 961–971 (2012). Article CAS PubMed Google Scholar * Cecconi, C., Shank, E.A., Bustamante, C. & Marqusee, S.
Direct observation of the three-state folding of a single protein molecule. _Science_ 309, 2057–2060 (2005). Article CAS PubMed Google Scholar * Shank, E.A., Cecconi, C., Dill, J.W.,
Marqusee, S. & Bustamante, C. The folding cooperativity of a protein is controlled by its chain topology. _Nature_ 465, 637–640 (2010). Article CAS PubMed PubMed Central Google
Scholar * Faham, S. & Bowie, J.U. Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. _J. Mol. Biol._ 316, 1–6
(2002). Article CAS PubMed Google Scholar * Joh, N.H. et al. Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins. _Nature_ 453, 1266–1270 (2008).
CAS PubMed Google Scholar * Dürr, U.H., Gildenberg, M. & Ramamoorthy, A. The magic of bicelles lights up membrane protein structure. _Chem. Rev._ 112, 6054–6074 (2012). Article
PubMed PubMed Central CAS Google Scholar * Wang, Y.C., Zhang, Y.J. & Ha, Y. Crystal structure of a rhomboid family intramembrane protease. _Nature_ 444, 179–183 (2006). Article CAS
PubMed Google Scholar * Wu, Z.R. et al. Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. _Nat. Struct. Mol. Biol._ 13,
1084–1091 (2006). Article CAS PubMed Google Scholar * Ben-Shem, A., Fass, D. & Bibi, E. Structural basis for intramembrane proteolysis by rhomboid serine proteases. _Proc. Natl.
Acad. Sci. USA_ 104, 462–466 (2007). Article CAS PubMed Google Scholar * Vinothkumar, K.R. Structure of rhomboid protease in a lipid environment. _J. Mol. Biol._ 407, 232–247 (2011).
Article CAS PubMed PubMed Central Google Scholar * Lemberg, M.K. & Freeman, M. Cutting proteins within lipid bilayers: rhomboid structure and mechanism. _Mol. Cell_ 28, 930–940
(2007). Article CAS PubMed Google Scholar * Freeman, M. Rhomboid proteases and their biological functions. _Annu. Rev. Genet._ 42, 191–210 (2008). Article CAS PubMed Google Scholar *
Ha, Y., Akiyama, Y. & Xue, Y. Structure and mechanism of rhomboid protease. _J. Biol. Chem._ 288, 15430–15436 (2013). Article CAS PubMed PubMed Central Google Scholar *
Vinothkumar, K.R. & Freeman, M. Intramembrane proteolysis by rhomboids: catalytic mechanisms and regulatory principles. _Curr. Opin. Struct. Biol._ 23, 851–858 (2013). Article CAS
PubMed Google Scholar * Lemberg, M.K. Sampling the membrane: function of rhomboid-family proteins. _Trends Cell Biol._ 23, 210–217 (2013). Article CAS PubMed Google Scholar * Baker,
R.P. & Urban, S. Architectural and thermodynamic principles underlying intramembrane protease function. _Nat. Chem. Biol._ 8, 759–768 (2012). Article CAS PubMed PubMed Central Google
Scholar * Paslawski, W. et al. Cooperative folding of a polytopic α-helical membrane protein involves a compact N-terminal nucleus and nonnative loops. _Proc. Natl. Acad. Sci. USA_ 112,
7978–7983 (2015). Article CAS PubMed PubMed Central Google Scholar * Cecconi, C., Shank, E.A., Marqusee, S. & Bustamante, C. in _DNA Nanotechnology: Methods and Protocols_ 1st edn.
Vol. 749 (eds. Zuccheri, G. & Samorì, B.) 255–271 (Humana Press, 2011). * Min, D. et al. Mechanical unzipping and rezipping of a single SNARE complex reveals hysteresis as a
force-generating mechanism. _Nat. Commun._ 4, 1705–1714 (2013). Article PubMed CAS Google Scholar * Bae, W. et al. Programmed folding of DNA origami structures through single-molecule
force control. _Nat. Commun._ 5, 5654–5661 (2014). Article CAS PubMed Google Scholar * Gosse, C. & Croquette, V. Magnetic tweezers: micromanipulation and force measurement at the
molecular level. _Biophys. J._ 82, 3314–3329 (2002). Article CAS PubMed PubMed Central Google Scholar * Saleh, O.A., Allemand, J.F., Croquette, V. & Bensimon, D. Single-molecule
manipulation measurements of DNA transport proteins. _ChemPhysChem_ 6, 813–818 (2005). Article CAS PubMed Google Scholar * Kim, K. & Saleh, O.A. A high-resolution magnetic tweezer
for single-molecule measurements. _Nucleic Acids Res._ 37, 136–142 (2009). Article CAS Google Scholar * Lipfert, J., Hao, X.M. & Dekker, N.H. Quantitative modeling and optimization of
magnetic tweezers. _Biophys. J._ 96, 5040–5049 (2009). Article CAS PubMed PubMed Central Google Scholar * De Vlaminck, I. & Dekker, C. Recent advances in magnetic tweezers. _Annu.
Rev. Biophys._ 41, 453–472 (2012). Article CAS PubMed Google Scholar * Greenleaf, W.J., Frieda, K.L., Foster, D.A.N., Woodside, M.T. & Block, S.M. Direct observation of hierarchical
folding in single riboswitch aptamers. _Science_ 319, 630–633 (2008). Article CAS PubMed PubMed Central Google Scholar * Alegre-Cebollada, J., Kosuri, P., Rivas-Pardo, J.A. &
Fernandez, J.M. Direct observation of disulfide isomerization in a single protein. _Nat. Chem._ 3, 882–887 (2011). Article CAS PubMed PubMed Central Google Scholar * Stigler, J.,
Ziegler, F., Gieseke, A., Gebhardt, J.C.M. & Rief, M. The complex folding network of single calmodulin molecules. _Science_ 334, 512–516 (2011). Article CAS PubMed Google Scholar *
Carrion-Vazquez, M. et al. Mechanical and chemical unfolding of a single protein: A comparison. _Proc. Natl. Acad. Sci. USA_ 96, 3694–3699 (1999). Article CAS PubMed PubMed Central
Google Scholar * Liphardt, J., Onoa, B., Smith, S.B., Tinoco, I. Jr. & Bustamante, C. Reversible unfolding of single RNA molecules by mechanical force. _Science_ 292, 733–737 (2001).
Article CAS PubMed Google Scholar * Beaugrand, M. et al. Lipid concentration and molar ratio boundaries for the use of isotropic bicelles. _Langmuir_ 30, 6162–6170 (2014). Article CAS
PubMed PubMed Central Google Scholar * Curnow, P. & Booth, P.J. Combined kinetic and thermodynamic analysis of α-helical membrane protein unfolding. _Proc. Natl. Acad. Sci. USA_ 104,
18970–18975 (2007). Article CAS PubMed PubMed Central Google Scholar * Otzen, D.E. Mapping the folding pathway of the transmembrane protein DsbB by protein engineering. _Protein Eng.
Des. Sel._ 24, 139–149 (2011). Article CAS PubMed Google Scholar * Watters, A.L. et al. The highly cooperative folding of small naturally occurring proteins is likely the result of
natural selection. _Cell_ 128, 613–624 (2007). Article CAS PubMed Google Scholar * Cymer, F., von Heijne, G. & White, S.H. Mechanisms of integral membrane protein insertion and
folding. _J. Mol. Biol._ 427, 999–1022 (2015). Article CAS PubMed Google Scholar * Kim, S.J. & Skach, W.R. Mechanisms of CFTR folding at the endoplasmic reticulum. _Front.
Pharmacol._ 3, 201 (2012). CAS PubMed PubMed Central Google Scholar * Curnow, P. et al. Stable folding core in the folding transition state of an α-helical integral membrane protein.
_Proc. Natl. Acad. Sci. USA_ 108, 14133–14138 (2011). Article CAS PubMed PubMed Central Google Scholar * Jefferson, R.E., Blois, T.M. & Bowie, J.U. Membrane proteins can have high
kinetic stability. _J. Am. Chem. Soc._ 135, 15183–15190 (2013). Article CAS PubMed Google Scholar * Otzen, D.E. Folding of DsbB in mixed micelles: a kinetic analysis of the stability of
a bacterial membrane protein. _J. Mol. Biol._ 330, 641–649 (2003). Article CAS PubMed Google Scholar * Kim, B.L., Schafer, N.P. & Wolynes, P.G. Predictive energy landscapes for
folding α-helical transmembrane proteins. _Proc. Natl. Acad. Sci. USA_ 111, 11031–11036 (2014). Article CAS PubMed PubMed Central Google Scholar * Urban, S. & Wolfe, M.S.
Reconstitution of intramembrane proteolysis in vitro reveals that pure rhomboid is sufficient for catalysis and specificity. _Proc. Natl. Acad. Sci. USA_ 102, 1883–1888 (2005). Article CAS
PubMed PubMed Central Google Scholar * Faham, S., Ujwal, R., Abramson, J. & Bowie, J.U. in _Membrane Protein Crystallization_ 1st edn. Vol. 63 (eds. DeLucas, L.J.) 109–125 (Academic
Press, 2009). * Ujwal, R. & Bowie, J.U. Crystallizing membrane proteins using lipidic bicelles. _Methods_ 55, 337–341 (2011). Article CAS PubMed PubMed Central Google Scholar *
Whitelegge, J.P. et al. Toward the bilayer proteome, electrospray ionization-mass spectrometry of large, intact transmembrane proteins. _Proc. Natl. Acad. Sci. USA_ 96, 10695–10698 (1999).
Article CAS PubMed PubMed Central Google Scholar * Kessler, D.A. & Rabin, Y. Distribution functions for filaments under tension. _J. Chem. Phys._ 121, 1155–1164 (2004). Article CAS
PubMed Google Scholar * Schwaiger, I., Sattler, C., Hostetter, D.R. & Rief, M. The myosin coiled-coil is a truly elastic protein structure. _Nat. Mater._ 1, 232–235 (2002). Article
CAS PubMed Google Scholar * Schuler, B., Lipman, E.A. & Eaton, W.A. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. _Nature_ 419,
743–747 (2002). Article CAS PubMed Google Scholar * Yang, W.Y. & Gruebele, M. Folding at the speed limit. _Nature_ 423, 193–197 (2003). Article CAS PubMed Google Scholar *
Rhoades, E., Cohen, M., Schuler, B. & Haran, G. Two-state folding observed in individual protein molecules. _J. Am. Chem. Soc._ 126, 14686–14687 (2004). Article CAS PubMed Google
Scholar * Kubelka, J., Hofrichter, J. & Eaton, W.A. The protein folding 'speed limit'. _Curr. Opin. Struct. Biol._ 14, 76–88 (2004). Article CAS PubMed Google Scholar *
Chung, H.S., Louis, J.M. & Eaton, W.A. Experimental determination of upper bound for transition path times in protein folding from single-molecule photon-by-photon trajectories. _Proc.
Natl. Acad. Sci. USA_ 106, 11837–11844 (2009). Article CAS PubMed PubMed Central Google Scholar * Gebhardt, J.C.M., Bornschlogla, T. & Rief, M. Full distance-resolved folding energy
landscape of one single protein molecule. _Proc. Natl. Acad. Sci. USA_ 107, 2013–2018 (2010). Article CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS This
work was supported by the National Creative Research Initiative Program (Center for Single-Molecule Systems Biology to T.-Y.Y.) funded by the National Research Foundation of Korea and
Marine Biotechnology Program (20150220 to T.-Y.Y.) funded by the Ministry of Oceans and Fisheries of Korea, and supported by US National Institutes of Health grant 2R01GM063919 to J.U.B.
AUTHOR INFORMATION Author notes * Duyoung Min Present address: Present address: Department of Chemistry and Biochemistry, University of California–Los Angeles, Los Angeles, California, USA.,
* Duyoung Min and Robert E Jefferson: These authors contributed equally to this work. AUTHORS AND AFFILIATIONS * National Creative Research Initiative Center for Single-Molecule Systems
Biology, KAIST, Daejeon, South Korea Duyoung Min & Tae-Young Yoon * Department of Physics, KAIST, Daejeon, South Korea Duyoung Min & Tae-Young Yoon * Department of Chemistry and
Biochemistry, University of California–Los Angeles, Los Angeles, California, USA Robert E Jefferson & James U Bowie Authors * Duyoung Min View author publications You can also search for
this author inPubMed Google Scholar * Robert E Jefferson View author publications You can also search for this author inPubMed Google Scholar * James U Bowie View author publications You
can also search for this author inPubMed Google Scholar * Tae-Young Yoon View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS D.M., R.E.J.,
J.U.B. and T.-Y.Y. designed the experiments. R.E.J. expressed and purified proteins. D.M. prepared the DNA-protein hybrid sample and performed the magnetic tweezers experiments. All of the
authors analyzed the data and contributed to writing of the manuscript. CORRESPONDING AUTHORS Correspondence to James U Bowie or Tae-Young Yoon. ETHICS DECLARATIONS COMPETING INTERESTS The
authors declare no competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY TEXT AND FIGURES Supplementary Results, Supplementary Figures 1–10 and Supplementary Tables 1–3.
(PDF 2696 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Min, D., Jefferson, R., Bowie, J. _et al._ Mapping the energy landscape for second-stage
folding of a single membrane protein. _Nat Chem Biol_ 11, 981–987 (2015). https://doi.org/10.1038/nchembio.1939 Download citation * Received: 13 July 2015 * Accepted: 14 September 2015 *
Published: 19 October 2015 * Issue Date: December 2015 * DOI: https://doi.org/10.1038/nchembio.1939 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this
content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative