Play all audios:
ABSTRACT Symbiotic nitrogen fixation (SNF) in legume-rhizobia serves as a sustainable source of nitrogen (N) in agriculture. However, the addition of inorganic N fertilizers significantly
inhibits SNF, and the underlying mechanisms remain not-well understood. Here, we report that inorganic N disrupts iron (Fe) homeostasis in soybean nodules, leading to a decrease in SNF
efficiency. This disruption is attributed to the inhibition of the Fe transporter genes _Natural Resistance-Associated Macrophage Protein 2a and 2b_ (_GmNRAMP2a&2b)_ by inorganic N.
GmNRAMP2a&2b are predominantly localized at the tonoplast of uninfected nodule tissues, affecting Fe transfer to infected cells and consequently, modulating SNF efficiency. In addition,
we identified a pair of N-signal regulators, nitrogen-regulated GARP-type transcription factors 1a and 1b (GmNIGT1a&1b), that negatively regulate the expression of _GmNRAMP2a&2b_,
which establishes a link between N signaling and Fe homeostasis in nodules. Our findings reveal a plausible mechanism by which soybean adjusts SNF efficiency through Fe allocation in
response to fluctuating inorganic N conditions, offering valuable insights for optimizing N and Fe management in legume-based agricultural systems. SIMILAR CONTENT BEING VIEWED BY OTHERS IMA
PEPTIDES REGULATE ROOT NODULATION AND NITROGEN HOMEOSTASIS BY PROVIDING IRON ACCORDING TO INTERNAL NITROGEN STATUS Article Open access 29 January 2024 TRANSFER CELLS MEDIATE NITRATE UPTAKE
TO CONTROL ROOT NODULE SYMBIOSIS Article 08 June 2020 THE NAC TRANSCRIPTION FACTORS SNAP1/2/3/4 ARE CENTRAL REGULATORS MEDIATING HIGH NITROGEN RESPONSES IN MATURE NODULES OF SOYBEAN Article
Open access 05 August 2023 INTRODUCTION Legumes have a natural ability to fix atmospheric nitrogen (N2) into organic form through their N-fixing symbiosis system, making them a major
sustainable source of nitrogen for agriculture1. However, despite providing ~50 million tons of N per annum into agricultural systems, symbiotic nitrogen fixation (SNF) by legumes still
falls short of the amount provided by inorganic N fertilizers2. Excessive use of N fertilizers not only incurs environmental and economic costs, but also inhibits SNF in legumes3.
Understanding the mechanisms underlying this inhibition can help develop strategies to balance nodule-based N fixation with soil N fertilization. The nutrient exchange between legumes and
rhizobia is a mutually beneficial and essential aspect of their symbiosis, where rhizobia provide fixed N to legumes in exchange for carbohydrates and mineral elements. This exchange takes
place in root nodules of legumes4. One of the mineral elements provided by host plants is iron (Fe), which is indispensable for SNF in rhizobia5. Root nodules of legumes typically contain a
higher concentration of Fe than other vegetative organs. At the maturity stage, more than 40% of Fe accumulates in soybean nodules6. The large amount of Fe is used as cofactors and
components of the proteins (nitrogenase, leghemoglobin, ferredoxin, etc.) that are essential for SNF5. However, Fe deficiency can severely inhibit nodule formation and development, and
consequently, SNF efficiency7,8, especially for legumes grown in calcareous soils where Fe becomes poorly soluble9. Vacuolar Fe plays an important role in maintaining Fe homeostasis in
plants. Plants can sequester excess Fe to the vacuoles, preventing its toxic accumulation. When Fe is deficient, vacuolar Fe can be mobilized and transported to other parts of the plant,
providing a readily available source of Fe for metabolic processes10,11. This movement of Fe is often mediated by Fe transport proteins at the tonoplast. In Arabidopsis, Fe can be
compartmented in vacuoles of embryos by VIT1 and mobilized by AtNRAMP3&4 during seed germination12. Similarly, legume nodules rely on a complex system of Fe transporters to maintain Fe
homeostasis, with numerous transport family members, including NRAMP, VIT, YSL, ZIP, and MATE, working together to move Fe from the vasculature to the infected cells, and finally to the
basic nitrogen-fixing unit, symbiosome1,5,13. Despite the advancements made in nodule Fe transport, it remains unclear if nodules have specialized Fe-storing cells and relevant Fe
transporters. Legumes have an auto-regulation of nodulation (AON) system that responds to external N sources and fine-tunes nodulation to prevent carbon loss when N is abundant1. During the
stages of SNF, metabolic and transport processes associated with SNF are precisely regulated to adapt to external N14. High levels of inorganic N can cause premature aging of the nodules,
thus terminating the transfer of nutrients from the host plant to the nodules15. Transcription factors, Nodule Inception (NIN), and NIN-like proteins (NLP) play crucial roles in both
nodulation and SNF in legumes1,16,17. These N-responsive NIN / NLPs modulate signaling pathways responsible for nodule formation and development18,19. Recent studies have found that NLP2 and
NIN can directly activate the expression of leghemoglobin, an oxygen-binding phytoglobin that carries heme (an Fe-containing molecule) in nodules of _Medicago truncatula_16. However, the
mechanism by which inorganic N regulates nodule Fe homeostasis is still largely unknown. In this study, we found that the addition of inorganic N significantly disrupts the Fe homeostasis in
soybean nodules, thereby affecting SNF. This process is attributed to the inhibition of the expression of the Fe transporter genes _GmNRAMP2a&2b_ by inorganic N. A previous report
suggested that GmNRAMP2b (also known as GmDMT1) facilitated Fe transport from the cytoplasm of infected cells into symbiosomes20, functioning in a similar manner to VIT-like (VTL)
transporters21,22,23. However, our findings reveal that both GmNRAMP2a&2b were Fe influx transporters localized at the tonoplast of uninfected tissues of nodule. They act in a manner of
genetic compensation, to facilitate Fe transfer from uninfected to infected cells, which are indispensable for SNF in soybean nodules. We further demonstrate that the expression of
_GmNRAMP2a&2b_ is negatively regulated by two Nitrate-Inducible GARP-type Transcriptional Repressor (NIGT) family members, which reveals a regulatory network for N-dependent fine-tuning
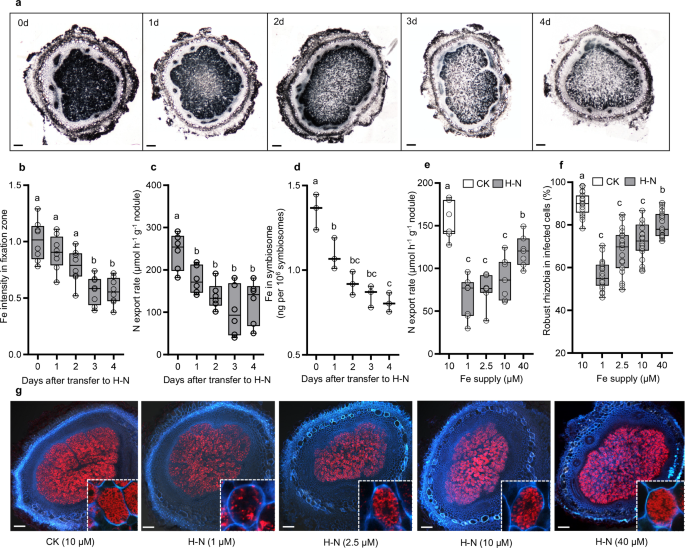
of Fe transport in symbiotic systems. RESULTS INORGANIC N INHIBITS SNF BY DISRUPTING FE HOMEOSTASIS To investigate the effects of inorganic N on nodule Fe homeostasis, we conducted Perls/DAB
staining on nodules exposed to high levels of inorganic N (H–N). Our observations revealed that Fe accumulated predominantly in the nitrogen fixation zones of the nodules. Furthermore, with
prolonged N supply, the Fe signal gradually diminished (Fig. 1a, b), which was also accompanied by a reduction in N export rate in nodules (Fig. 1c). Consistently, Fe content in symbiosome
was gradually decreased after transfer to H–N (Fig. 1d), suggesting that inorganic N blocks Fe delivery and disrupts Fe balance within the nodules. Subsequently, we assessed the inhibitory
effects of N on SNF under varying Fe concentrations. By analyzing nodule N export rates and the proportion of RFP-expressing rhizobia within infected cells, we observed that high Fe supply
mitigated the inhibitory effects of N on SNF, while low Fe resulted in the opposite outcome (Fig. 1e–g). These findings collectively suggest that disruption of Fe homeostasis may be one of
the pathways through which inorganic N inhibits SNF in nodules. EXPRESSION OF _GMNRAMP2A&2B_ IN NODULES IS RESPONSIVE TO EXTERNAL N AND FE LEVELS To investigate the molecular basis on
N-regulated Fe homeostasis in nodules, we first carried out comparative RNA-seq analysis of mature nodules and screened out 142 genes responding to both Fe deficiency (-Fe) and H-N (Fig. 2a,
Supplementary Data 1). Among these genes, we classified Fe homeostasis-related genes into four clusters based on their response patterns (Fig. 2b, c). Intriguingly, two _NRAMP_ genes were
both down-regulated by H-N and up-regulated by -Fe (Fig. 2c). A phylogenetic tree of NRAMPs from rice, soybean and Arabidopsis was constructed and the result suggested that these two
_NRAMPs_ are the most similar homologues in the soybean genome (Supplementary Fig. 1a). They are 98% identical at the amino acid level and very conserved in their trans-membrane domains
(Supplementary Fig. 1b), suggesting that they may play complementary roles in their biological functions. Combined with the previous report that they are a pair of paralogs resulting from
genome duplication events24, we therefore named them _GmNRAMP2a_ and _GmNRAMP2b_ hereinafter. _GmNRAMP2a_ and _2b_ are expressed in most tissues according to the Phytozome database, and only
_GmNRAMP2b_ is highly expressed in nodules (Fig. 2d). The expression of these two genes in other tissues may play a role similar to that of AtNRAMP3 and 4 in Arabidopsis25. Real-time RT-PCR
showed that _GmNRAMP2b_ was primarily expressed in the fixation zone of nodules (Fig. 2e). _GmNRAMP2b_ exhibited much higher expression levels than _GmNRAMP2a_ (Fig. 2e). The expression of
_GmNRAMP2b_ but not _GmNRAMP2a_ was gradually increased with the days after inoculation, reaching its peak at 21 days and subsequently declining gradually (Fig. 2f). Consistent with RNA-seq
data, both genes were up-regulated by -Fe and down-regulated by H–N, but were not varied by other nutrient deficiencies, such as Mg, Mo, S, Mn or Zn (Fig. 2g). Notably, under both -Fe and
H–N conditions, the expression of _GmNRAMP2a_ and _2b_ remained suppressed (Fig. 2h, i), suggesting that N signals play a dominant role in expression regulation. Furthermore, regardless of
whether the N source is ammonium or nitrate, both can trigger the suppression of gene expression (Supplementary Fig. 2a, b). GMNRAMP2A&2B ARE TONOPLAST-TARGETED PROTEINS IN NODULE
UNINFECTED TISSUES To verify their tissue and cell specificities, we generated transgenic hairy roots carrying green fluorescent protein (GFP) driven by _GmNRAMP2a_ or _2b_ promoters. In
situ immunostaining results from nodules at 17 and 30 dpi showed that GFP antibody (anti-GFP) signals were observed mainly in fixation zone of _pGmNRAMP2a:GFP_ transgenic nodules under
Fe-deficient conditions, whereas no signals could be observed under Fe-sufficient conditions (Fig. 3a, Supplementary Fig. 3a). By contrast, anti-GFP signals can be detected in
_pGmNRAMP2b:GFP_ transgenic nodules under both Fe-deficient and -sufficient conditions (Fig. 3b, Supplementary Fig. 3b). Using an RFP-expressing rhizobium strain as a marker for infected
cells, we found that both genes were expressed in those cells that were not colonized by rhizobia (Fig. 3c, d, Supplementary Fig. 3c, d), indicating the uninfected cell-specific expression
of _GmNRAMP2a&2b_. The signal intensity from both transgenic nodules showed a -Fe-induced pattern, and followed the order: Fixation zone > Vasculature > Cortex (Fig. 3e,
Supplementary Fig. 3e). Furthermore, Fe limitation did not alter uninfected cell-specific expression of _GmNRAMP2b_ (Fig. 3f, Supplementary Fig. 3f). We next examined the subcellular
localization of GmNRAMP2a&2b in tobacco (_Nicotiana tabacum_) leaf protoplasts. Fluorescence signals from both GmNRAMP2a-GFP and GmNRAMP2b-GFP proteins were observed at the tonoplast,
which were easily distinguishable from signals of the plasma membrane (PM) marker FM4-64 FX and chlorophyll autofluorescence (Fig. 3g, h). We subsequently investigated the subcellular
localizations of GmNRAMP2a&2b in yeast, and found both of them localized specifically to vacuolar membrane (Supplementary Fig. 4a). To further verify their tissue and subcellular
localizations in soybean nodules, we generated transgenic hairy roots carrying _pGmNRAMP2a/2b: GmNRAMP2a/2b-GFP_, and inoculated them with rhizobium. To obtain a more clearly visible signal,
we treated _pGmNRAMP2a: GmNRAMP2a-GFP_ transgenic nodules with Fe deficiency. In situ immunostaining results from nodules at 17 and 30 dpi showed that in both transgenic nodules, anti-GFP
signals mainly located in uninfected cells (smaller cell size) of fixation zone that was in close proximity to infected cells (larger cell size; Fig. 3i–j, Supplementary Fig. 3g-h). They
were also observed in the pericycle of nodule vascular tissues (Supplementary Fig. 5a, b). Furthermore, these signals showed a ring-like structure inside the cell but outside the nucleus
(Fig. 3i–j, Supplementary Figs. 3g, h, 5c, d), suggesting that GmNRAMP2a&2b are targeted to the cell tonoplast. We next examined their protein levels under H–N supply, and found that
both proteins (GmNRAMP2b under +Fe and GmRNAMP2a under -Fe conditions) were very susceptible to H–N supply, with a rapid decrease of anti-GFP signal abundances in nodules after being
transferred to H–N for 12 and 24 h (Fig. 3k–l, Supplementary Figs. 5e, f). GmNRAMP2b was previously identified on the symbiosome membrane (SM) via immunoelectron microscopy20. We also used
this method with GFP-specific antibodies for confirmation. However, we found that GmNRAMP2b-GFP was not detected on the SM but specifically located on the tonoplast of uninfected cells
(Supplementary Fig. 6a). This subcellular localization was further confirmed by western blot analysis, which showed that the GmNRAMP2b protein tagged with GFP displayed the same
fractionation pattern as the V-type ATPase (a known tonoplast membrane marker protein), but differed from the pattern shown by H+-ATPase (a known plasma membrane marker protein).
Furthermore, we did not detect the GFP-tagged GmNRAMP2b protein in any subcellular structure of the symbiosomes, whereas the Nodulin-26 protein (a marker protein for the SM) showed a
specific localization on the SM (Supplementary Fig. 6b). These results indicate that GmNRAMP2b has a specific localization to the tonoplast of uninfected cells. YEAST COMPLEMENTATION TEST OF
GMNRAMP2A&2B To investigate the Fe transport activity, we transformed the ORFs of _GmNRAMP2a_, _GmNRAMP2b_, _GmVTL1a_21, and the full-length (FL) cDNA of _GmNRAMP2b_ individually into
yeast WT strain BY4741 or _Δccc1_ mutant26, and then isolated vacuoles for Fe determination. The purity of extracted vacuoles was qualified by vacuolar marker ALP (Fig. 4a) and integrity was
examined by FM4-64 staining (Supplementary Fig. 4b). We found the vacuolar Fe content in _GmVTL1a_ transformants was remarkably higher than vector control after gene induction by galactose
for 1 or 2 h. In contrast, the Fe content in _GmNRAMP2a&2b_ and _GmNRAMP2b-FL_ transformants was much lower than vector control in BY4741 strains (Supplemental Fig. 4c). Fe accumulation
in vacuoles was always lower in _Δccc1_ mutant compared to BY4741, due to defects in vacuolar Fe storage of _Δccc1_ (Fig. 4b). However, _GmVTL1a_ and _GmNRAMP2a&2b_ transformants in
_Δccc1_ mutant still exhibited the same trends as observed in BY4741 (Fig. 4b). To further elucidate their potential Fe transport abilities, we fused the PM signal peptide ENO2(169)27 in
front of the ORF of _GmNRAMP2a_ or _2b_, and transformed them into _fet3fet4_ strain. The ENO2(169)-fused GmNRAMP2a or 2b showed both PM and tonoplast positioning (Supplementary Fig. 4d).
Although low-Fe supply makes _fet3fet4_ strain grow poorly, transformation with either _ENO2_(_169_)-_GmNRAMP2a_ or _2b_ promoted the growth of _fet3fet4_ (Fig. 4c, d). In parallel,
short-term 57Fe uptake experiment showed that Fe uptake in both _ENO2_(_169_)-_GmNRAMP2a&2b_ transformants was significantly higher than the vector control (Fig. 4e). Taken together,
these results indicate that GmNRAMP2a&2b are Fe influx transporters and mediate the transport of Fe from the vacuoles to the cytoplasm in yeast cells. _GMNRAMP2A&2B_ MODULATE SNF
ACTIVITY BY AFFECTING FE TRANSFER TO INFECTED CELLS Due to the dominant expression of _GmNRAMP2b_ (Fig. 2e, f), we first generated a _nramp2b_ single mutant by CRISPR-Cas9 for phenotype
analysis (Supplementary Fig. 7a). Regardless of Fe availability, there were no differences in plant growth or nodule development, as well as in the response of SNF activity to H–N conditions
(Supplementary Fig. 8a–g), except that the expression of _GmNRAMP2a_ in the nodule of _nramp2b_ mutant was significantly increased (Supplementary Fig. 8h). We therefore generated two
double-knockout lines named _nramp2ab-1_ and _nramp2ab-2_ by CRISPR-Cas9 (Supplementary Fig. 7b, c). Phenotypic analysis showed that there was no difference in plant growth between WT and
two double-knockout lines under non-symbiotic conditions (non-inoculation with H–N supply). However, after inoculated with rhizobium, the growth of two double-knockout lines was well below
that of WT (Fig. 5a, b). Meanwhile, compared to WT plants, the double-knockout lines exhibited a significant decrease of 41% in single nodule weight (Fig. 5c, d), 48% in N export rate (Fig.
5e), and 46% in SNF activity (Supplementary Fig. 9a). Furthermore, the exogenous addition of high-Fe fully restored the nodule weight and SNF activity in double-knockout lines (Supplementary
Fig. 9b, c), suggesting that the phenotypic defects in mutants are due to Fe limitation. Notably, _GmNRAMP2a&2b_ knockout did not alter rhizobia invasion, nodule primordium initiation
or nodule number per plant (Supplementary Fig. 9d–h). These results reveal that GmNRAMP2a&2b may not participate in the early-stage processes of rhizobia infection or nodule
organogenesis, but rather affecting the later-stage processes of nodule development and SNF. We next determined Fe status in nodules, and found that in the _nramp2ab_ mutants, Fe intensity
in fixation zone, as well as the Fe concentration in symbiosome were significantly decreased compared to the WT (Fig. 5f, g, Supplementary Fig. 9i). Subsequently, we isolated the intact
uninfected and infected cell protoplasts through cell wall digestion and microcapillary separation (Supplementary Fig. 9j). Our results showed that Fe in the uninfected cell of _nramp2ab_
mutants increased by 112%, while it decreased by 25% in the infected cell (Supplementary Fig. 9k), suggesting that GmNRAMP2a&2b are helpful for Fe delivery from uninfected cell to
infected cell. To investigate whether GmNRAMP2a&2b affect the delivery of other trace elements in nodules, LA-ICP-TOF technology was used to observe the accumulation of trace elements in
nodules. We found that the differences in Cu, Mn and Zn accumulation between WT and mutant nodules were not significant as compared to Fe (Supplementary Fig. 9l). Phenotypic analysis of WT
and mutants under various micronutrient deficiency conditions revealed that the mutants exhibited unaltered biomass of nodule and seedling under Cu, Mn, or Zn-deficient conditions
(Supplementary Fig. 9m, n). These findings indicate that GmNRAMP2a&2b play a more significant role in delivering Fe to nodules compared to other trace elements. We used ferritin as a
marker for intracellular Fe levels10 and found decreased ferritin in _nramp2ab_ mutants through transcriptomic and western blot assays (Supplementary Figs. 10, 11a; Supplementary Data 2),
suggesting that defective Fe transport from vacuoles reduces cytoplasmic Fe. Conversely, _vtl1_ mutants21 showed increased ferritin (Supplementary Figs. 10, 11a; Supplementary Data 2),
suggesting disrupted Fe transport to symbiosomes and resultant cytoplasmic Fe accumulation. In parallel, both _nramp2ab_ and _vtl1_ mutants showed reduced nitrogenase NifH and leghemoglobins
(Lbs) levels (Supplementary Figs. 10, 11a, Supplementary Data 2), indicating that disruption of Fe homeostasis (whether a deficiency or an excess) impair nodule N fixation. We also examined
how mutations affected the expression of gene families associated with Fe transport (_NRAMP_, _VIT_, _YSL_, _ZIP_, and _OPT_). The _nramp2ab_ mutation led to decreased expression of three
_NRAMP_ genes, two _VIT_ genes, one _YSL_ gene, and two _ZIP_ genes. In contrast, the _vtl1_ mutation resulted in the reduced expression of two _NRAMP_ genes, two _VIT_ genes, and five _ZIP_
genes, while also increasing the expression of two _NRAMP_ genes, two _VIT_ genes, and three _YSL_ genes (including _GmYSL7_28,29), one _ZIP_ gene, and one _OPT_ gene (Supplementary Fig.
10). Overall, the effects of the _vtl1_ mutation on the expression of Fe transporters are more pronounced than the _nramp2ab_ mutation in nodules. To further validate these result, we
expressed _GmNRAMP2b-GFP_ in _vtl1_ mutants, and found that GmNRAMP2b-GFP was undetectable in _vtl1_ mutants (Supplementary Fig. 11b). These results suggest that Fe homeostasis in nodules
relies on the complex interplay and coordination of multiple transporters. GMNRAMP2A&2B ARE INVOLVED IN THE INHIBITION OF SNF BY INORGANIC N Due to the inhibitory effect of H–N on
_GmNRAMP2a&2b_ expression, we hypothesized that H–N might impact nodule Fe homeostasis through the regulation of GmNRAMP2a&2b. To test this hypothesis, we investigated the phenotypes
of _nramp2ab_ mutants under H–N conditions. We observed 38% and 35% inhibitions by H–N in N export rate in the WT and the mutants, respectively. Meanwhile, the WT showed a 33% reduction in
Fe accumulation in H–N, whereas the mutants displayed only 16% reduction (Fig. 5e–g). H–N led to evident Fe bodies in uninfected cells of the WT, along with the appearance of lytic vacuoles
in infected cells (Fig. 5h–j). In contrast, these Fe bodies were widespread in the mutants regardless of N levels, and H–N resulted in an increase in the number and size of lytic vacuoles
compared to the WT (Fig. 5h-j). To further verify this hypothesis, we constructed overexpression lines and found that in both _GmNRAMP2a_ and _GmNRAMP2a&2b_ overexpression lines,
although the plant seedlings and nodule phenotypes showed no significant changes under symbiotic conditions (Supplementary Fig. 12), H–N supply to the nodules led to significantly higher N
export rate and Fe accumulation in fixation zone compared to the WT (Fig. 5k-l), while Fe bodies in uninfected cells and lytic vacuoles in infected cells was significantly lower than in the
WT (Fig. 5m, n). Taken together, these results indicate that the inhibitory effect of H–N on SNF depends on GmNRAMP2a&2b. GMNIGT1A&1B NEGATIVELY REGULATE N-RESPONSIVE EXPRESSION OF
_GMNRAMP2A&2B_ To investigate _cis_-acting elements responsive to N in the _GmNRAMP2b_ promoter, we first constructed five vectors expressing _GUS_ driven by different _GmNRAMP2b_
promoter segments (P1-P5, Supplementary Fig. 13a). The transgenic nodules carrying each vector showed much lower expression of _GmNRAMP2b_ after H–N supply for 1 d (Supplementary Fig. 13b),
suggesting that −500 bp promoter region is enough for its N-responsive expression. Based on the known N-responsive _cis_-acting elements19,30,31, we found three NIGT and one NIN-like
_cis_-acting elements (named NIE and NRE respectively) in both −500 bp promoters of _GmNRAMP2a&2b_ (Supplementary Fig. 13c, d). Considering the great importance of NIN/NLPs in root
nodules, we first investigated their regulatory role on _GmNRAMP2a&2b_ expression. Soybean has four NINs functioning redundantly in nodulation, and ten NLPs with unknown functions32.
Since NINs and NLPs in soybean exist in the manner of paralogous gene pairs (Supplementary Fig. 14a), we selected one of each pair and overexpressed them in nodules for further
investigation. Among these transgenic events, only overexpressing _GmNIN1b_ reduced the expression of _GmNRAMP2a&2b_ as well as single nodule weight (Supplementary Fig. 14b–d). However,
GmNIN1b could bind to neither promoter of _GmNRAMP2a_ nor _2b_ by yeast one-hybrid assay (Supplementary Fig. 14e). Both GmNIN1b and AtNLP133 could bind to 4×NREAtNIR1 but not to
4×NREGmNRAMP2a/2b, suggesting that the predicted NREGmNRAMP2a/2b is not the true NIN/NLP binding site, and the reduced expression of _GmNRAMP2a&2b_ might be an indirect effect due to the
inhibited nodulation by _GmNIN1b_ overexpression30,32. We next investigated the regulatory role of NIGT proteins on _GmNRAMP2a&2b_ expression. There are four NIGTs (two paralogous gene
pairs) in soybean with highest sequence homology to AtNIGTs in Arabidopsis (Supplementary Fig. 15a), but only one pair of them (named _GmNIGT1a&1b_) are dominantly expressed in nodules
(Fig. 6a). Transcriptomic data revealed that similar to _GmNRAMP2a&2b_, both _GmNIGT1a_&_1b_ were specifically expressed in nodules, and exhibited much higher expression levels in
fixation zone (Fig. 6b). Furthermore, both _GmNIGT1a_&_1b_ were only up-regulated by H–N and had no response to Fe depletion (Fig. 6c). Their expression significantly increased in
response to H–N from 7 to 50 dpi (Supplementary Fig. 2c, d), suggesting the role of GmNIGT1a&1b in N perception is active throughout early to late nodule development. This remarkable
similarity in gene expression pattern indicates a potential complementary relationship of _GmNIGT1a_ and _1b_ in their biological functions. We therefore selected _GmNIGT1a&1b_ for yeast
one-hybrid assay, and found that both of them fused with transcriptional activation domain (AD) of GAL4 can bind to the promoters of _GmNRAMP2a&2b_, as well as the NIGT cis-enriched
sequence (6 × NIE, Fig. 6d). To verify whether GmNIGT1a&1b have the same repression effects on gene transcription as AtNIGTs, we employed transcriptional repressor activity assays in
both tobacco leaves and soybean nodules. The effector (GAL4 binding domain (BD)-fusion protein) is able to bind with the GAL4 cis-enriched sequence (6×GAL4) upstream of the reporter gene
(_GFP_), and thereby affects _GFP_ expression if the effector has a transcriptional activation/repression activity. The results revealed that in both systems (tobacco leaves and soybean
nodules), expression of _GmNIGT1a_, _GmNIGT1b_ or _AtNIGT1.1_ individually led to a significant reduction in the expression of reporter gene (Fig. 6e–g). This indicates that GmNIGT1a&1b
may function as transcriptional repressors in soybean. On the other hand, EMSA result showed that both GmNIGT1a and 1b could directly bind to all five of putative _cis_-acting elements in
vitro, although their binding affinity to probe 5 was markedly weaker (Fig. 6h, Supplementary Fig. 13c). In situ immunostaining results showed that both GmNIGT1a and 1b were localized in all
nodule cells except for the infected cell, with high-intensity signals at nuclei and weak signals at cytoplasm (Fig. 6i, Supplementary Fig. 15b–d). GMNIGT1A&1B REGULATE FE HOMEOSTASIS
IN NODULES To clarify the role of GmNIGT1a&1b in nodule Fe homeostasis and SNF, we constructed the double knockout line (_nigt1ab-cr_) by CRISPR-Cas9 (Supplementary Fig. 7d), as well as
the knockdown (_nigt1ab-RNAi_) and overexpression lines (_GmNIGT1a-OE_). There was no change in the growth of seedlings and nodules in the _nigt1ab-RNAi_ and _nigt1ab-cr_ lines, while
_GmNIGT1a-OE_ lines displayed notable decreases in seedling and nodule growth (Fig. 7a–d). Knockdown or knockout of _GmNIGT1a_&_1b_ alleviated the inhibition of _GmNRAMP2a&2b_
expression by H–N, while overexpressing _GmNIGT1a_ resulted in a low-level expression of _GmNRAMP2a&2b_ regardless of N availability (Fig. 7e–f). When inorganic N was supplied to the
nodules, _nigt1ab-RNAi_ and _nigt1ab-cr_ lines exhibited higher N export rates and Fe accumulation in nodules than WT (Fig. 7g-j). In contrast, _GmNIGT1a-OE_ lines displayed notable
reductions in N export rates and Fe accumulation in fixation zone (Fig. 7g–j). In parallel, the number of Fe bodies in uninfected cells and lytic vacuoles in infected cells was significantly
lower in _nigt1ab-RNAi_ and _nigt1ab-cr_ lines but higher in _GmNIGT1a-OE_ lines (Fig. 7k–m). _p35S_-driven overexpression is not limited to nodules and can potentially lead to indirect
effects on nodule development. To achieve in situ overexpression, we utilized the _GmNIGT1a_ promoter to drive the expression of _GmNRAMP2b_ and the _GmNRAMP2b_ promoter to drive _GmNIGT1a_
expression in soybean hairy roots. As expected, nodules with _pGmNIGT1a:GmNRAMP2b-GFP_ exhibited a stronger GFP signal under H–N conditions, and the Fe accumulation and distribution in these
nodules followed a trend similar to nodules with _p35S:GmNRAMP2b_. Conversely, nodules containing _pGmNRAMP2b:GmNIGT1a-GFP_ showed a stronger GFP signal in control conditions, and the Fe
accumulation and distribution mirrored the pattern seen in nodules with _p35S:GmNIGT1a_ (Supplementary Fig. 16). Taken together, these observations indicate a direct and specific regulatory
role of NIGT1-NRAMP2 module in Fe homeostasis and N-inhibited SNF within nodules. To explore more downstream targets of GmNIGT1a&1b, we conducted a comparative transcriptomic assay,
identifying 47 genes potentially regulated by GmNIGT1a&1b (Supplementary Data 3). These genes are up-regulated in _nigt1ab_ double knockout line, but down-regulated in _GmNIGT1a-OE_ line
and under H–N conditions (Supplementary Fig. 17a). Notably, our findings reveal that, in addition to _GmNRAMP2a&2b_, putative Fe homeostasis-related genes such as _IRON MAN_ (_IMA_s)
and _BRUTUS_s are also under the regulation of GmNIGT1a&1b (Supplementary Fig. 17b). Whether they are involved in Fe homeostasis requires further study. DISCUSSION Recent reports have
revealed that VTL transporters facilitate Fe transport into symbiosomes for SNF in legume nodules21,22,23. Although previously GmNRAMP2b, also termed GmDMT1, was presumed to act as an Fe
transporter at the SM of soybean nodules20, this has been brought into question by researchers which suggest it is unlikely to be involved in exporting Fe out of cells34,35. Given that the
physiological role of GmNRAMP2b in SNF remains unconfirmed, its contribution to Fe homeostasis within nodules was not yet understood. Furthermore, GmNRAMP2b and GmVTL1a showed different
transcriptional response to N and Fe availability (21; Fig. 2g–i), suggesting their distinct physiological functions. In this study, we cannot verify the previously reported results that
GmNRAMP2b locates at the SM of infected cell20. Instead, we demonstrate that GmNRAMP2b and its paralog GmNRAMP2a works primarily in uninfected tissues (Fig. 3a–f, Supplementary Fig. 3a–f),
and whether expressed in yeast, tobacco protoplasts or soybean nodules, GmNRAMP2a&2b were localized at the tonoplast (Fig. 3g–j, Supplementary Fig. 4a). Besides, analyses of two
single-cell databases (zhailab.bio.sustech.edu.cn/single cell soybean; soybeancellatlas.org) consistently indicate that GmNRAMP2a&2b and GmNIGT1a&1b are predominantly found in the
uninfected cells of soybean nodules. Moreover, upon investigating the published soybean SM proteomics data36, we found no detection of GmNRAMP2a, 2b, GmNIGT1a, or 1b proteins. Nevertheless,
_GmNRAMP2b_ was still expressed at a low level in infected cells (Fig. 3d–f), suggesting its potential minor role in these cells. GmNRAMP2b was considered as a Fe influx transporter due to
its ability of rescuing the growth of the _fet3fet4_ mutant defective in ferrous Fe uptake20. However, GmNRAMP2b was found to be non-PM localized (Supplementary Fig. 4a), which led us to
reason that complementation of _fet3fet4_ strain by GmNRAMP2b was probably caused by an indirect effect. The yeast system does have some limitations when used to express exogenous proteins,
such as the mislocalization of plant chloroplast membrane proteins to the yeast cell membrane37. However, it is a commonly used system for gene functional complementation tests38. Therefore,
by ectopically expressing _GmNRAMP2a&2b_ on the yeast plasma membrane, we were able to successfully complement the _fet3fet4_ mutant (Fig. 4c–e), suggesting that GmNRAMP2a&2b act as
transporters for ferrous Fe influx, and in planta, they facilitate the transport of Fe from the vacuole to the cytoplasm. In soybean nodules, uninfected cells play an important role in
supporting nitrogen fixation. However, little attention has been paid to these uninfected cells, and it was previously thought that they were only involved in C and N metabolism39,40. In
this study, we observed that compared to WT plants, _nramp2ab_ mutants accumulated less Fe in infected cells and symbiosomes, but more Fe in uninfected cells (Fig. 5f–i, Supplementary Fig.
9i–k). This suggests that GmNRAMP2a&2b can mobilize Fe from the vacuoles of uninfected cells, which can then be transported to the apoplast and subsequently enter infected cells via
other Fe transporters (41, Fig. 8). Alternatively, the mobilized Fe could be transported via plasmodesmata through the symplastic pathway, as suggested by previous research (42, Fig. 8).
Infected cells in soybean nodules are mostly occupied by rhizobia, and have no vacuoles for nutrient storage, while uninfected cells have large central vacuoles and provide a large surface
area for interaction with the infected tissue43. Therefore, GmNRAMP2a&2b in uninfected cells can ensure timely and dynamic release of Fe from vacuoles to infected cells for SNF. While
_nramp2ab_ mutants exhibited a reduced nodule size and nitrogenase activity, their phenotype is not as severe as for _vtl1_ mutants, which show little activity for SNF21. We reason that in
addition to GmNRAMP2a&2b-involved Fe transport, other Fe transport pathways and relevant transporters are also important for Fe delivery to infected cells (Supplementary Fig. 10). For
example, a NRAMP homolog from _Medicago truncatula_ is localized at the PM of infected cells, responsible for transport of Fe from the apoplastic into infected cells41. The equivalent
transporter in soybean may collaborate with NRAMP2 to ensure Fe homeostasis in nodules. In general, VTL1 and NRAMP2 proteins play different roles in soybean nodules. VTL1 is an indispensable
protein for maintaining the basic N fixation of infected cells, while NRAMP2 predominantly functions as a regulator in uninfected cells, with its role becoming more pronounced when the
nodule has a higher demand for Fe. Organisms have developed genetic robustness to maintain normal development in response to harmful mutations. In addition to gene redundancy, genetic
compensation response is recently suggested as another important mechanism for genetic robustness, where one or more paralogs are upregulated to substitute for the compromised activity of
another44,45. In this study, we found that the paralogs _GmNIGT1a_ and _1b_ exhibited highly similar expression patterns (Fig. 6a–c), suggesting they may have complementary functions. In
contrast, although GmNRAMP2a&2b shared the same Fe transport activities and subcellular localizations (Figs. 3, 4), they differed in expression patterns. Compared to the dominant
expression of _GmNRAMP2b_, _GmNRAMP2a_ was low-level expressed and upregulated only under conditions of Fe-depletion or sole _GmNRAMP2b_ knockout (Fig. 2f, g, Supplementary Fig. 8 h). This
suggests that GmNRAMP2a&2b possess an asymmetrically redundant role in Fe transport. Instead, GmNRAMP2a has evolved to provide an active dosage compensation when large amounts of Fe are
required in nodules. As soybean is an ancient tetraploid with ~75% of current genes present in multiple copies, and _GmNRAMP2a&2b_ are paralogs resulting from genome duplication
events24, our study offers functional evidence supporting the notion that genome duplication enhances soybean’s environmental adaptability. It was found that both _GmNRAMP2a&2b_ were
regulated by N and Fe availability in nodules (Fig. 2). For Fe-regulation, it seems reasonable because these two genes encode Fe transporters, and similar regulation of _NRAMP_ genes by Fe
availability as well as their regulatory networks have been widely reported in other species46. For N-regulation, it seems reasonable particularly for nodule organs as Fe is indispensable
for SNF, and SNF is tightly controlled by external inorganic N levels. However, N-modulated Fe homeostasis is little understood. Intriguingly, our study revealed a NIGT-NRAMP regulatory
module in nodules. NIGT family proteins are a group of G2-like GARP-type transcription factors that were previously shown to suppress expression of a series of genes related to nitrate
transport and assimilation, and are thereby recognized as N-satiation-signal transducers to prevent excessive N accumulation and energy consumption47. The existence of a NIGT-NRAMP
regulatory module suggests that a novel function obtained by soybean NIGTs in the symbiotic system, is to regulate Fe transport to achieve dynamic Fe supply for SNF. How NIGT perceive and
transmit N signals within nodules remains unknown. However, it is interesting to note that most NIGT1s are identified as direct targets of NAC transcription factors48. This suggests that
NACs could at least partially mediate the transmission of N signals through the NIGT1 signaling pathways. Meanwhile, the genes downstream of NIGT1 include not only _NRAMP2_ but also _IMAs_
and _BRUTUSs_ (Supplementary Fig. 17b). IMA peptides positively regulate Fe homeostasis in plants by interacting with the E3 ubiquitin ligase BRUTUS, which is required for the degradation of
transcription factors involved in the Fe deficiency response49,50. In root nodules of _Lotus japonicus_, IMA peptides have recently been reported to regulate nitrogen fixation51, which
highlights the essential role of IMA-mediated Fe provision in regulating N-related physiological processes. Whether IMA peptides regulate NRAMP2 or act independently to maintain Fe balance
in root nodules requires further study. Nodule formation and nitrogen fixation require a lot of energy, and legumes have developed strategies to adjust nodule numbers and SNF levels in
response to changes in N levels in the environment18. As Fe is vital for both the host and rhizobia, legumes may save resources by preventing Fe allocation to SNF via NIGT perception of N
signals when sufficient N is available to plants. Therefore, in agricultural fertilization management, it is essential to emphasize the supplementation of the trace element - Fe, to mitigate
the inhibition of nodule SNF by inorganic nitrogen fertilizers. METHODS PLANT MATERIAL AND GROWTH CONDITIONS The stable gene knockout mutants were obtained using CRISPR-Cas9 technology in
the soybean (_Glycine max_) genotype Williams 8252. The guide RNA sequence for each mutant is shown in Supplementary Data 4. Transgenic seedlings were then generated through _Agrobacterium
tumefaciens_ (EHA105)-mediated transformation53. The predicted editing sites in T1 seedlings were sequenced and those with frameshift mutations were selected. T2 homozygous seeds were
collected for phenotypic analysis. To generate stable _GmNIGT1a&1b_ knockdown material (_nigt1ab-RNAi_), a 273-bp (position 622-894 starting from ATG) conserved region of
_GmNIGT1a&1b_ with 96% nucleotide identity, was amplified and inserted into the _Asc_I and _Swa_I sites of pFGC5941 in the sense orientation. This construct was then inserted into the
_Xba_I and _BamH_I sites in the anti-sense orientation. 35S promoter was used for the RNAi construction. To construct the stable overexpression lines, the ORFs of _GmNRAMP2a_, _GmNRAMP2b_,
and _GmNIGT1a_ were amplified and individually inserted into the _Asc_I and _Xba_I sites of pFGC5941-_p35S_ construct. 35S promoter was used for the overexpression line construction.
Transgenic seedlings were then generated through _Agrobacterium tumefaciens_ (EHA105)-mediated transformation. _GmNRAMP2a_&_2b_ double overexpression lines were obtained by
co-transformation. Soybean seeds were surface-sterilized by exposure to chlorine gas overnight prior to germinating in sterilized vermiculite. After 4 d, seedlings were inoculated with
_Bradyrhizobium_ strain BXYD3 or RFP-expressing strain21, and cultured with a low-N nutrient solution in vermiculite. Seedlings were then transplanted and cultivated in a low-N nutrient
solution before various Fe or N treatments. Low-N solution was prepared with 1/10 of the N in the base nutrient solution (5.3 mM)54. High-N solution was supplemented with NH4NO3 to achieve a
total N concentration of 20 mM. EDTA-Fe (10 μM) was used for plant culture if not otherwise specified. To generate transgenic soybean composite plants, the hypocotyl injection method for
hairy root transformation was utilized according to ref. 21. The transformed hairy roots from 25-d-old seedlings were inoculated with _Bradyrhizobium_ strain and cultured in a low-N nutrient
solution before Fe or N treatment. All seedlings were grown in a growth chamber with a 13 h/26 °C day and 11 h/24 °C night regime, with daytime light provided by light-emitting diode at an
intensity of 400 μmol photons m−2s−1, and relative humidity maintained at 65%. Nutrient solutions were renewed every 2 days and pH was adjusted to 5.8. Roots were continuously aerated
through an air pump. PERLS/DAB STAINING Nodule samples were embedded in resin according to the method of ref. 55. Briefly, Nodules were incubated overnight in fixation solution containing
50% (v/v) ethanol, 5% (v/v) glacial acetic acid, and 10% (v/v) formaldehyde solution and vacuum infiltrated for 30 minutes. The fixed nodules were dehydrated in a series of 50%, 60%, 70%,
80%, and 90% ethanol solutions, and then overnight dehydrated in 100% ethanol. Samples were embedded in Technovit 7100 resin (Kulzer) according to the kit instructions, and thin sections (7
µm) were prepared. These sections were vacuum infiltrated for 15 min each with equal volumes of 4% (v/v) HCl and 4% (w/v) K-ferrocyanide (Perls stain solution), and incubated for 30 min at
room temperature. For DAB intensification, fixed sections were washed with distilled water and incubated in a methanol solution containing 10 mM NaN3 and 0.3% (v/v) H2O2 for 1 h. After
washing with PBS, sections were then incubated in an intensification solution containing 0.025% (w/v) DAB, 0.005% (v/v) H2O2, 0.005% (w/v) CoCl2 and 0.1 M PBS (pH 7.4) for 30 min prior to
being washed with distilled water to stop the reaction. These sections were photographed using an optical microscope (Nikon Ni-U, Japan). ISOLATION OF INTACT SYMBIOSOMES Intact symbiosomes
were isolated according to ref. 21. Briefly, fresh nodules were ground gently in an ice-cold homogenizing buffer. Samples were then filtered through 4 layers of miracloth (Millipore, USA),
and slowly transferred onto the top of a 30/60% (v/v) Percoll gradient solution. After centrifuging at 4000 g for 15 min, symbiosomes were collected from the 60% Percoll fraction (including
the 30/60% interface). Collected symbiosomes were rinsed three times with a wash buffer. The number of symbiosomes was counted using a hemocytometer under a light microscope (Primo Star,
Carl Zeiss, Germany). Samples were then digested in concentrated nitric acid for measurement of Fe concentrations using ICP-MS (Agilent 7900, USA). N EXPORT RATE AND ACETYLENE REDUCTION
ASSAY Basal regions of soybean shoots (2 cm above the roots) were excised with a razor, and then xylem sap was collected for 1 h, and the concentration of ureides was determined using
colorimetric analysis of glyoxylate derivatives according to the ref. 54. N export rate of nodule was calculated as the total ureide content divided by the fresh weight of nodules. Acetylene
reduction activity in nodules was determined according to ref. 54. Briefly, nodules were isolated and kept in an air tight glass bottle, and then immediately exposed to acetylene gas for 2
h. After injecting 0.5 M NaOH to terminate the reaction, a 0.3 mL gas sample was extracted and injected into a gas chromatograph (GC-2014, SHIMADZU, Japan) for ethylene determination.
TRANSCRIPTOMIC ANALYSIS For the transcriptomic analysis depicted in Fig. 2a–c, the -Fe treatment involved transplanting seedlings at 10 dpi into a low-N and Fe-free solution for 7 days. The
H–N treatment entailed initially transplanting seedlings at 10 dpi into a low-N solution and EDTA-Fe ( + Fe) for 6 days, followed by exposure to H–N solution for 1 day. The CK treatment
involved transplanting seedlings at 10 dpi into a low-N and +Fe solution for 7 d. For the transcriptomic analysis depicted in Supplementary Fig. 10, the -Fe treatment involved transplanting
seedlings at 14 dpi from WT, _nramp2ab_, and _vtl1_ mutants into a low-N and Fe-free solution for 7 days. The H–N treatment involved transplanting seedlings at 20 dpi into a H–N solution for
1 d. Low-N and +Fe treatments were used as CK. For the transcriptomic analysis depicted in Supplementary Fig. 17, nodules grown in a low-N solution at 21 dpi from WT, _nigt1ab-cr_ and
_GmNIGT1a-OE_ lines were used for RNA-seq analysis. The H–N treatment involved transplanting seedlings of WT at 20 dpi into a H–N solution for 1 d. Nodule samples were harvested and quickly
frozen by liquid nitrogen for subsequent RNA sequencing analysis using an Illumina HiSeqTM 2500 platform (Novogene, China). Genes with fold change larger than 2 (or log2 FC > 1) were
selected. QUANTITATIVE GENE EXPRESSION ANALYSIS To investigate the gene expression in nodules, nodules at 21dpi were separated into three parts for RNA extraction: nodule conjugated root
segments with nodules removed, nodule cortex, and fixation zone21. For time-course analysis, nodules grown in a low-N solution were harvested for RNA extraction at 7, 14, 17, 21, 30, 40, and
50 dpi. To investigate the expression response to various nutrient stresses, nodules grown in a low-N solution at 10 dpi were transferred to a low-N and Fe-, Mg-, Mo-, Mn-, Zn- or S-free
solution for 7 d, or nodules grown in a low-N solution at 16 dpi were treated with high-N for 1 d, and then were harvested for RNA extraction. To investigate the expression response to N and
Fe interaction, nodules grown in a low-N solution at 21dpi were treated with H–N, -Fe or a combination of both for 1, 2, 3 or 4 d. To investigate the expression response to different N
source, nodules grown in a low-N solution at 21dpi were treated with 10 mM ammonium, 10 mM nitrate or a combination of both for 1, 2, 3 or 4 d. For real-time reverse transcription (RT)-PCR,
total RNA was extracted using _TransZol_ Up Plus RNA Kit (TransGen, China). 500 ng of RNA was used for complementary DNA (cDNA) synthesis using TransScript One-Step genomic DNA Removal and
cDNA Synthesis Super Mix (TransGen, China). Gene expression levels were determined by real-time RT-PCR using TransStart Top Green qPCR SuperMix (TransGen, China). The housekeeping gene EF-1a
was used as an internal control. Normalized relative expression was calculated by the ΔΔCt method. The primers used for RT-PCR are shown in Supplementary Data 4. PHYLOGENETIC ANALYSIS
Protein sequences were obtained from Phytozome (phytozome-next.jgi.doe.gov/) and miyakogusa.jp (kazusa.or.jp/lotus) database. The alignment analysis of protein sequences was performed using
MEGA7. TISSUE AND SUBCELLULAR LOCALIZATION To investigate tissue-specific expression of _GmNRAMP2a&b_, their respective 2.5 kb promoter sequences were amplified and cloned into the
pFGC5941_-GFP_ vector to create the _pGmNRAMP2a:GFP_ and _pGmNRAMP2b:GFP_ constructs. To determine subcellular localization of GmNRAMP2a&b proteins in nodules, the ORFs of
_GmNRAMP2a&b_ were individually amplified and inserted into the above constructed vectors to create the _pGmNRAMP2a:GmNRAMP2a-GFP_ and _pGmNRAMP2b: GmNRAMP2b-GFP_ constructs. To
determine subcellular localization of GmNIGT1a&1b proteins in nodules, sequences including 2.5 kb upstream promoter and genomic gene sequence were amplified and cloned into the
pFGC5941_-GFP_ vector to create the _pGmNIGT1a: GmNIGT1a-GFP_ and _pGmNIGT1b: GmNIGT1b-GFP_ constructs. The primers are shown in Supplementary Data 4. These constructs were transformed into
_Agrobacterium rhizogenes_ strain K599 for hairy-root transformation. The transformed hairy roots from 25-d-old seedlings were inoculated with rhizobia and cultured in a low-N nutrient
solution before Fe or N treatment. Nodules at 17dpi or 30 dpi were collected for the immunostaining. Immunostaining was performed according to the methods of Liu et al. 21. A polyclonal
anti-GFP (1:1000; Thermo Scientific, USA) was used for the primary antibody. Alexa Fluor 488 or 555 goat anti-rabbit IgG (1:2000; Thermo Scientific, USA) were used for the secondary
antibody. Calcofluorwhite (1:2000; Sigma, USA) and DAPI (1:500; Solarbio, China) were used for cell wall and nucleus staining, respectively. To investigate the subcellular localization of
GmNRAMP2a&2b in tobacco (_Nicotiana tabacum_) protoplasts, the ORFs of both genes were amplified and then inserted into pFGC5941-_p35S-GFP_ to obtain _p35S:GmNRAMP2a-GFP_ and
_p35S:GmNRAMP2b-GFP_. FM4-64 FX (Thermo Scientific, USA) was used as a PM marker. The protoplasts used for transient expression analysis were extracted from tobacco grown in Fe-sufficient
conditions and transformed by the polyethylene glycol (PEG) method56. To investigate the subcellular localization of GmNRAMP2a&b in yeast, _GmNRAMP2a-GFP_ and _GmNRAMP2b-GFP_ sequences
were amplified and cloned into pYES2 vector (V82520, Invitrogen, USA) respectively. Subsequently, the PM signal peptide ENO2(169)27 was amplified from yeast DNA and inserted in front of
_GmNRAMP2a/2_b-GFP. The primers are shown in Supplementary Data 4. The reconstructed vectors were transformed into wild-type strain BY4741 using the S.c.easy Comp Transformation Kit
(Invitrogen, USA). Fluorescence was observed with a confocal scanning microscope (LSM880, Carl Zeiss, Germany) after yeast growth with galactose. IMMUNOELECTRON MICROSCOPY The nodules
samples were fixed with 4% paraformaldehyde (PFA) in phosphate buffer (PB, 0.1 M, pH 7.0) for 30 min followed by agar embedding and oscillating slicing. The oscillating sections (120-150 μm)
were rapidly frozen and fixed in a high pressure freezing apparatus (Wohlwend Compact 03, Wohlwend, Switzerland), and then transferred to 0.2% uranyl acetate in pure acetone at −90°C for
subsequent freeze substitution in a freeze substitution instrument (EM AFS2, Leica, Germany). Then the frozen water in the samples was gradually replaced by acetone and resin Lowicryl HM20
(Electron Microscopy Sciences, USA) at −45 °C. Embedding and UV polymerization were performed stepwise at −40 °C. For Immunoelectron microscopy, the ultrathin sections were immunolabled with
anti-GFP antibody (1:50, Abcam, UK) as primary antibody for 90 min, followed by treatment with goat anti-rabbit IgG conjugated with 15-nm-diameter gold particles as secondary antibody
(1:100, Abcam, UK) for 60 min. Sections were then stained and observed using TEM (HT7800, Hitachi, Japan). WESTERN BLOT ANALYSIS For western blot of nitrogenase, ferritin and leghemoglobin,
nodules from WT, _nramp2ab_ and _vtl1_ mutants were harvested and ground into powder in liquid N. Sample was loaded equally onto an SDS-PAGE gel, and then blotted to a polyvinylidene
fluoride membrane (Immobilon-P, Millipore, USA). The membrane was probed with anti-NifH (1:2000; Agrisera, Sweden), anti-ferritin (1:2500; Agrisera, Sweden), or anti-leghemoglobin57,
anti-actin (1:5000; ABclonal, China) overnight, and followed with their corresponding horseradish peroxidase (HRP)-conjugated second antibodies (anti-chicken IgY (1:10,000; Thermo
Scientific, USA) for NifH, anti-rabbit IgG (1:5000; Biosharp, China) for ferritin; anti-Goat IgG (1:1000; Solarbio, China) for leghemoglobin, anti-mouse IgG (1:5000; TransGen, China) for
actin) for 1 h. For western blot of yeast marker proteins, anti-ALP (1:1000; Abcam, UK), anti-PGK (1:2000; Abcam, UK) and anti-porin (1:1000; Abcam, UK) were used as primary antibodies.
Anti-mouse (for ALP and porin) and anti-rabbit (for PGK) IgG HRP-conjugated antibody (1:2000; TransGen, China) were used as second antibodies. The HRP signals were detected using the
SuperSignal West Dura Trial Kit (Thermo Scientific, USA) with an Amersham Imager 600 (GE Healthcare Bio-Sciences AB, Sweden). For full scan blots, please see the Source Data file. For
western blot of nodules’ membrane proteins, intact symbiosomes were isolated as described above. Subsequently, symbiosomes were separated into symbiosome membrane (SM), symbiosome space (SS)
and bacteroids (B) according to ref. 58. The supernatant from percoll gradient centrifugation were further fractionated using discontinuous sucrose gradients (20–60%) according to ref. 36.
Immunoblot analysis was performed using primary antibodies for GFP (1:1000; TransGen, China), V-type ATPase (1:2000; Agrisera Sweden), H+-ATPase (1:2000; Agrisera Sweden), Nodulin-26 (1:500;
the synthetic peptide TKNTSETIQRSDSLV was used to immunize rabbits to obtain antibodies against Nodulin-26). Anti-mouse (for GFP) and anti-rabbit (for V-type ATPase, H+-ATPase, and
Nodulin-26) IgG HRP-conjugated antibody (1:2000; TransGen, China) were used as second antibodies. COMPLEMENTATION TEST IN YEAST The amplified ORFs of _GmNRAMP2a_, _GmNRAMP2b_, _GmVTL1a_ or
full-length cDNA of _GmNRAMP2b_ were cloned into pYES2 vector, which was then transformed into BY4741 (WT) or _Δccc1_ yeast strain26 using the S.c.easy Comp Transformation Kit (Thermo
Scientific, USA). The primers are shown in Supplementary Data 4. Yeast transformants were selected on synthetic defined medium without Ura (SD-Ura) containing 2% glucose. After liquid
culture with glucose to exponential phase, yeast transformants were incubated with SD-Ura containing 2% galactose, 1% raffinose and 1 mM FeSO4 for 0, 0.5, 1 or 2 h. Vacuoles were isolated
according to the methods of Li et al.26 with the following modifications: 300 ml of yeast cells was collected by centrifugation at 3000 g for 3 min. The cells were resuspended in 10 ml of
0.1 M Tris-HCl (pH 9.3) and 10 mM dithiothreitol, and incubated for 10 min at 30 °C. The cells were washed once with spheroplast buffer (1.2 M sorbitol, 20 mM potassium phosphate, pH 7.4)
and incubated with 500 U/ml lyticase (Solarbio, China) for 2 h at 30 °C. Spheroplasts were collected by centrifugation at 3500 g for 5 min and resuspended in 3.5 ml of 15% ficoll buffer (15%
ficoll, 0.2 M sorbitol, 10 mM PIPES-KOH, pH 6.8). 3.5 μl of DEAE-Dextran (50 mg/ml) was added to the spheroplasts, and the sample was incubated for 3 min on ice and then for 5 min at 30 °C.
3.5 μl of MgCl2 (1.5 M) was added to the lysate to terminate the reaction. The lysate was transferred to 13PA tubes (Koki Holdings, Japan) and overlaid with 3 ml of 8% Ficoll, 4 ml of 4%
Ficoll, and 1 ml of 0% Ficoll. The tubes were centrifuged at 110,000 g for 90 min. The vacuolar fraction was collected from the 0/4% interphase, and protein concentrations from vacuoles were
determined by a BCA protein assay reagent kit (TransGen, China). Samples were digested by concentrated nitric acid for Fe determination by ICP-MS (Agilent 7900, USA). To generate
PM-targeted proteins, the PM signal peptide ENO2(169) was amplified from yeast DNA and inserted into _pYES2-GmNRAMP2a/2b_ vectors. The primers are shown in Supplementary Data 4. The
recombinant vectors _pYES2-ENO2(169)-GmNRAMP2a/2b_ were transformed into Fe uptake defective mutant _fet3fet4_59. After selected by SD-Ura with glucose, yeast cells were cultured by YNB
(-Fe) medium with yeast synthetic Drop-out medium supplements (-Ura) and glucose to exponential phase, and then were spotted onto SD-Ura plates with galactose and different concentrations of
FeCl3. For yeast cell density determination, yeast cell suspensions were diluted to an OD600 of 0.1, and then incubated with galactose and different concentrations of FeCl3 at 30 °C for 21
h. The values of OD600 were dynamically determined. For short-term 57Fe uptake, yeast cells were cultured by YNB (-Fe) medium with yeast synthetic Drop-out medium supplements (-Ura) and
galactose to exponential phase, and collected by centrifugation at 3000 g for 5 min. The cells were washed twice with sterile water and incubated with 1, 5, 10 or 100 µM 57FeCl2 (96.1% 57Fe;
Trace Sciences International, Canada) for 5 min at RT. Yeast cells were collected and digested by concentrated nitric acid for 57Fe determination by ICP-MS using stable isotope mode
(Agilent 7900, USA). SEPARATION OF INFECTED AND UNINFECTED CELLS Intact infected and uninfected cell was isolated according to ref. 60 with some modifications. The fixation zone of mature
root nodules (1-2 g) was dug out and cut into pieces, and then incubated in 5 mL enzyme solution (1% cellulase R-10, 0.1% pectolyase Y-23, 0.6 M mannitol,10 mM MES-KOH (pH 5.7), 1 mM MgCl2,
0.5% BSA, 0.5% dextran sulfate) at 28°C for 30 min with gentle shaking (40 rpm). Samples were filtered through three-layer tea bag to remove small tissue debris and bacteroids. The residues
on the tea bag were then collected and washed three time with the same solution without enzyme. The cleaned samples were further incubated in 10 mL enzyme solution at 28°C without shaking
for 2 h, and followed with occasional shaking for 1 h. The samples were filtered by three-layer tea bag, and followed by 30 μm nylon mesh. Cells on the mesh were suspended in 20 mM MOPS-KOH
(pH 7.5) containing 0.6 M mannitol and 5 mM CaCl2 on ice. Infected cells (larger, irregular-shaped and reddish-brown color) and uninfected cells (smaller, regular-shaped and nearly
transparent) were separated by glass capillary tubes under a microscope. After quantified by using a hemocytometer, the collected cells were dried and digested by concentrated nitric acid
for Fe determination by ICP-MS (Agilent 7900, USA). LA-ICP-TOF-MS Nodules were first embedded in resin and then sliced into 10 µm thick sections according to the method mentioned in
Perls/DAB staining. These sections were analyzed using a LA unit (NWR 193ImageGEO; New Wave Research) with the following settings: energy: 1 J/cm2; scan rate: 16000 µm s−1; ablation
frequency: 200 Hz; spot size: 8\(\,\times \,\,\) µm. Element signals were obtained using TOF-ICP-MS (TOF-WERK, Switzerland) with the following settings: Vendor: Tofwerk; Type: icpTOF R;
Nebulizer gas flow: 1 L min−1; RF power: 1400 W; Detector: MCP; Dwell time: 5 ms. All element signals were normalized to 13C and converted to element images using iolite 4 software
(http://iolite-software.com/). Three biological replicates were tested. This experiment was performed by Shanghai Chemlabpro Technology Co., Ltd. TRANSCRIPTIONAL INHIBITION BY GMNIGTS OR
GMNINS/NLPS For segmental construction of _GmNRAMP2b_ promoters, the _GmNRAMP2b_ upstream regions of 2.5-kb, 2-kb, 1.5-kb, 1-kb, and 0.5-kb were amplified and cloned into the pFGC5941-GUS
vector containing _p35S: GFP_ cassette, respectively. The constructed vectors were transformed into hairy roots. Transgenic hairy roots from 25-d-old seedlings were inoculated with rhizobia
and grown in low-N solution for 17 d, and then treated with or without H-N solution for 1 d. Nodule samples were collected for RNA extraction and gene expression analysis. For
transcriptional repressor activity assays, _6×GAL4-TATA_ sequence was synthesized and inserted into pFGC5941-_p35S-GFP_ vector to construct the reporter plasmid. Each NIGT gene was amplified
and fused with GAL4 DNA binding domain (BD) at its N terminus to construct the effector plasmid. pFGC5941-_p35S-RFP_ vector was used as internal control. The reporter, effector and internal
control vectors were co-transformed into tobacco leaf, and GFP / RFP fluorescence signals were detected after 2-d incubation. In parallel, the reporter and effector were transformed into
hairy roots, and nodules at 17 dpi from hairy roots were used for gene expression analysis. To construct the overexpression lines, the ORFs of _GmNINs/GmNLPs_ were amplified and individually
inserted into the _Asc_I and _Xba_I sites of pFGC5941-_p35S_ construct. The constructed vectors were transformed into hairy roots, and nodules at 17 dpi from hairy roots were used for gene
expression and phenotypic analysis. YEAST ONE-HYBRID ASSAY For yeast one-hybrid assay, the ORFs of _GmNIGT1a&1b_, _GmNIN1b_ and _AtNLP1_ were amplified and cloned in frame after
transcriptional activation domain (AD) of GAL4 transcription factor in pB42AD respectively, which were used as effectors. The −500 bp promoter regions of _GmNRAMP2a&2b_, 4 × NREAtNIR1, 4
× NREGmNRAMP2a, 4 × NREGmNRAMP2b and 6 × NIE were cloned into the upstream of the _lacZ_ reporter gene in pLacZi vector respectively, which were used as reporters. The effectors combined
with the reporters were introduced into yeast strain EGY48 and cultured on SD medium (-Trp-Ura) containing X-gal at 30 °C. After 3 days, the yeast growth was photographed. EMSA ASSAY The
purified GST-NIGT1a/1b and the oligonucleotides described in Fig. 6h were used for EMSA. To perform the EMSA, Oligonucleotides were end labeled with or without (competitor) Cy5 as probes.
The coding sequence of NIGT1a/1b was individually introduced into PEGX4T-1. GST-NIGT1a/1b constructs and empty GST vectors were introduced into the _E. coli_ strain DE3 to induce protein
expression. The induced proteins were purified with Glutathione Sepharose 4B and then eluted with 10 mM glutathione. The Cy5-labelled probe (500 nM) was incubated with 2 μg recombinant
protein in a reaction (100 mM Tris-HCl [pH 7.5], 100 mM KCl, 50 mM MgCl2, 2.5 mM DTT) for 30 min at 4 °C. For competition assays, unlabeled double-stranded DNA was added to the binding
reaction. The EMSA reactions were subjected to electrophoresis on 3.5% polyacrylamide gels in 0.5×Tris borate EDTA (TBE) buffer at 4 °C in the dark. Electrophoresis was performed at 100 V
for 60 min. The fluorescence measurement of the polyacrylamide gel was detected on a LICOR Odyssey CLx system at 635 nm for excitation and 700 nm for emission. IN SITU OVEREXPRESSION OF
_GMNRAMP2B_ AND _GMNIGT1A_ To construct the overexpression lines, the 2.5 kb promoter sequences of _GmNramp2b_ with the ORF of _GmNIGT1a_ or the 2.4 kb promoter sequences of _GmNIGT1a_ with
the ORF of _GmNramp2b_ were amplified and individually inserted into the _EcoR_I and _Asc_I sites of pFGC5941-GFP construct. These constructs were transformed into _Agrobacterium rhizogenes_
strain K599 for hairy-root transformation. Transformed hairy roots from 25-d-old seedlings were inoculated with an RFP-tagged rhizobium. Transgenic nodules at 20 dpi were transplanted to
low-N (CK) or H–N for 2 d, and then were used for immunostaining and Perls/DAB staining. STATISTICS & REPRODUCIBILITY Statistical analyses were performed using GraphPad software. Means
were compared using One-way ANOVA (Tukey-test) or unpaired two-sided _t_-test. Sample sizes were chosen based on our experience on the experimental variability of this type of experiment and
the desire to get statistically significant data to support meaningful conclusions. The number of independent biological seedlings or replicates has been shown in each figure legend. No
data were excluded. Each experiment was repeated at least two times, and similar results were obtained. Seedlings were grown randomly in the growth chamber. Experiments were not blinded.
Data were always collected according to the genotype of plants. REPORTING SUMMARY Further information on research design is available in the Nature Portfolio Reporting Summary linked to this
article. DATA AVAILABILITY The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files. RNA-seq data have
been deposited at NCBI (National Center for Biotechnology Information, project accession number PRJNA875247). Source data are provided with this paper. REFERENCES * Roy, S. et al.
Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. _Plant Cell_ 32, 15–41 (2020). Article PubMed CAS Google Scholar * Canfield, D. E.,
Glazer, A. N. & Falkowski, P. G. The evolution and future of Earth’s nitrogen cycle. _Science_ 330, 192–196 (2010). Article ADS PubMed CAS Google Scholar * Sutton, M. A. et al. Too
much of a good thing. _Nature_ 472, 159–161 (2011). Article ADS PubMed CAS Google Scholar * Udvardi, M. & Poole, P. S. Transport and metabolism in legume-rhizobia symbioses. _Annu.
Rev. Plant Biol._ 64, 781–805 (2013). Article PubMed CAS Google Scholar * Brear, E. M., Day, D. A. & Smith, P. M. Iron: an essential micronutrient for the legume-rhizobium symbiosis.
_Front Plant Sci_. 4, 359 (2013). Article PubMed PubMed Central Google Scholar * Burton, J. W., Harlow, C. & Theil, E. C. Evidence for reutilization of nodule iron in soybean seed
development. _J. of Plant Nutr._ 21, 913–927 (1998). Article CAS Google Scholar * Slatni, T. et al. Metabolic changes of iron uptake in N2-fixing common bean nodules during iron
deficiency. _Plant Sci_. 181, 151–158 (2011). Article PubMed CAS Google Scholar * Slatni, T., Ben Salah, I., Kouas, S. & Abdelly, C. The role of nodules in the tolerance of common
bean to iron deficiency. _J. Plant Res._ 127, 455–465 (2014). Article PubMed CAS Google Scholar * Hansen, N. C., Schmitt, M. A., Anderson, J. E. & Strock, J. S. Iron deficiency of
soybean in the upper midwest and associated soil properties. _Agronomy J_ 95, 1595–1601 (2003). Article CAS Google Scholar * Briat, J. F., Curie, C. & Gaymard, F. Iron utilization and
metabolism in plants. _Curr. Opin. Plant Biol._ 10, 276–282 (2007). Article PubMed CAS Google Scholar * Curie, C. & Mari, S. New routes for plant iron mining. _New Phytol_. 214,
521–525 (2017). Article PubMed CAS Google Scholar * Conte, S. S. & Walker, E. L. Transporters contributing to iron trafficking in plants. _Mol. Plant_ 4, 464–476 (2011). Article
PubMed CAS Google Scholar * Banasiak, J., Jamruszka, T., Murray, J. D. & Jasinski, M. A roadmap of plant membrane transporters in arbuscular mycorrhizal and legume-rhizobium
symbioses. _Plant Physiol_. 187, 2071–2091 (2021). Article PubMed PubMed Central CAS Google Scholar * Streeter, J. & Wong, P. P. Inhibition of legume nodule formation and N2
fixation by nitrate. _Crit. Rev. Plant Sci._ 7, 1–23 (1988). Article CAS Google Scholar * Van de Velde, W. et al. Aging in legume symbiosis. a molecular view on nodule senescence in
_Medicago truncatula_. _Plant Physiol_. 141, 711–720 (2006). Article PubMed PubMed Central Google Scholar * Jiang, S. et al. NIN-like protein transcription factors regulate leghemoglobin
genes in legume nodules. _Science_ 374, 625–628 (2021). Article ADS PubMed CAS Google Scholar * Feng, J., Lee, T., Schiessl, K. & Oldroyd, G. E. D. Processing of NODULE INCEPTION
controls the transition to nitrogen fixation in root nodules. _Science_ 374, 629–632 (2021). Article ADS PubMed CAS Google Scholar * Lin, J. S. et al. NIN interacts with NLPs to mediate
nitrate inhibition of nodulation in _Medicago truncatula_. _Nat. Plants_ 4, 942–952 (2018). Article PubMed CAS Google Scholar * Nishida, H. et al. Different DNA-binding specificities of
NLP and NIN transcription factors underlie nitrate-induced control of root nodulation. _Plant Cell_ 33, 2340–2359 (2021). Article PubMed PubMed Central Google Scholar * Kaiser, B. N. et
al. The soybean NRAMP homologue, GmDMT1, is a symbiotic divalent metal transporter capable of ferrous iron transport. _Plant J_ 35, 295–304 (2003). Article PubMed CAS Google Scholar *
Liu, S. et al. A VIT-like transporter facilitates iron transport into nodule symbiosomes for nitrogen fixation in soybean. _New Phytol_. 226, 1413–1428 (2020). Article PubMed CAS Google
Scholar * Brear, E. M. et al. GmVTL1a is an iron transporter on the symbiosome membrane of soybean with an important role in nitrogen fixation. _New Phytol_. 228, 667–681 (2020). Article
PubMed CAS Google Scholar * Walton, J. H. et al. The _Medicago truncatula_ vacuolar iron transporter-like proteins VTL4 and VTL8 deliver iron to symbiotic bacteria at different stages of
the infection process. _New Phytol_. 228, 651–666 (2020). Article PubMed PubMed Central CAS Google Scholar * Qin, L. et al. Genome-wide identification and expression analysis of NRAMP
family genes in soybean (_Glycine Max_ L.). _Front Plant Sci_. 8, 1436 (2017). * Lanquar, V. et al. Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination
on low iron. _EMBO J._ 24, 4041–4051 (2005). Article PubMed PubMed Central CAS Google Scholar * Li, L., Chen, O. S., McVey Ward, D. & Kaplan, J. CCC1 is a transporter that mediates
vacuolar iron storage in yeast. _J. Biol. Chem._ 276, 29515–29519 (2001). Article PubMed CAS Google Scholar * Lopez-Villar, E. et al. Genetic and proteomic evidences support the
localization of yeast enolase in the cell surface. _Proteomics_ 6, S107–S118 (2006). Article PubMed Google Scholar * Wu, X. et al. GmYSL7 controls iron uptake, allocation, and cellular
response of nodules in soybean. _J. Integr. Plant Biol._ 65, 167–187 (2023). Article PubMed CAS Google Scholar * Gavrin, A. et al. Soybean Yellow Stripe-like 7 is a symbiosome membrane
peptide transporter important for nitrogen fixation. _Plant Physiol_. 186, 581–598 (2021). Article PubMed PubMed Central CAS Google Scholar * Wang, L. et al. A GmNINa-miR172c-NNC1
regulatory network coordinates the nodulation and autoregulation of nodulation pathways in soybean. _Mol. Plant_ 12, 1211–1226 (2019). Article PubMed CAS Google Scholar * Kiba, T. et al.
Repression of nitrogen starvation responses by members of the arabidopsis GARP-type transcription factor NIGT1/HRS1 subfamily. _Plant Cell_ 30, 925–945 (2018). Article PubMed PubMed
Central CAS Google Scholar * Fu, M., Sun, J., Li, X., Guan, Y. & Xie, F. Asymmetric redundancy of soybean Nodule Inception (NIN) genes in root nodule symbiosis. _Plant Physiol_. 188,
477–489 (2022). Article PubMed CAS Google Scholar * Konishi, M. & Yanagisawa, S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. _Nat. Commun._
4, 1617 (2013). Article ADS PubMed Google Scholar * González-Guerrero, M., Matthiadis, A., Sáez, Á. & Long, T. A. Fixating on metals: new insights into the role of metals in
nodulation and symbiotic nitrogen fixation. _Front Plant Sci_. 5, 45 (2014). Article PubMed PubMed Central Google Scholar * Clarke, V. C., Loughlin, P. C., Day, D. A. & Smith, P. M.
Transport processes of the legume symbiosome membrane. _Front Plant Sci_. 5, 699 (2014). Article PubMed PubMed Central Google Scholar * Luo, Y., Liu, W., Sun, J., Zhang, Z. R. &
Yang, W. C. Quantitative proteomics reveals key pathways in the symbiotic interface and the likely extracellular property of soybean symbiosome. _J. Genet. Genomics_ 50, 7–19 (2023). Article
PubMed CAS Google Scholar * Li, J. et al. Diel magnesium fluctuations in chloroplasts contribute to photosynthesis in rice. _Nat. Plants_ 6, 848–859 (2020). Article PubMed CAS Google
Scholar * Nielsen, J. Yeast systems biology: model organism and cell factory. _Biotechnol. J._ 14, e1800421 (2019). Article PubMed Google Scholar * Newcomb, E. H. & Tandon, S. R.
Uninfected cells of soybean root nodules: ultrastructure suggests key role in ureide production. _Science_ 212, 1394–1396 (1981). Article ADS PubMed CAS Google Scholar * White, J.,
Prell, J., James, E. K. & Poole, P. Nutrient sharing between symbionts. _Plant Physiol_. 144, 604–614 (2007). Article PubMed PubMed Central CAS Google Scholar * Tejada-Jimenez, M.
et al. _Medicago truncatula_ natural resistance-associated macrophage Protein1 is required for iron uptake by rhizobia-infected nodule cells. _Plant Physiol_. 168, 258–272 (2015). Article
PubMed PubMed Central CAS Google Scholar * Brown, S., Oparka, K., Sprent, J. & Walsh, K. B. Symplastic transport in the soybean nodules. _Soil Biol. Biochem._ 27, 387–399 (1995).
Article CAS Google Scholar * Selker, J. M. & Newcomb, E. H. Spatial relationships between uninfected and infected cells in root nodules of soybean. _Planta_ 165, 446–454 (1985).
Article PubMed CAS Google Scholar * Ma, Z. P. et al. PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. _Nature_ 568, 259–263 (2019). Article ADS
PubMed CAS Google Scholar * El-Brolosy, M. A. et al. Genetic compensation triggered by mutant mRNA degradation. _Nature_ 568, 193–197 (2019). Article ADS PubMed PubMed Central CAS
Google Scholar * Thomine, S. S., Schroeder, J. I. _Plant Metal Transporters With Homology To Proteins Of The NRAMP Family_, 113-121 (Landes Bioscience, Georgetown, Texas, 2004). * Ueda, Y.
& Yanagisawa, S. Perception, transduction, and integration of nitrogen and phosphorus nutritional signals in the transcriptional regulatory network in plants. _J. Exp. Bot._ 70,
3709–3717 (2019). Article PubMed CAS Google Scholar * Wang, X. et al. The NAC transcription factors SNAP1/2/3/4 are central regulators mediating high nitrogen responses in mature nodules
of soybean. _Nat. Commun._ 14, 4711 (2023). Article ADS PubMed PubMed Central CAS Google Scholar * Grillet, L., Lan, P., Li, W., Mokkapati, G. & Schmidt, W. IRON MAN is a
ubiquitous family of peptides that control iron transport in plants. _Nat. Plants_ 4, 953–963 (2018). Article PubMed CAS Google Scholar * Li, Y. et al. IRON MAN interacts with BRUTUS to
maintain iron homeostasis in Arabidopsis. _Proc. Natl. Acad. Sci. USA._ 118, e2109063118 (2021). Article PubMed PubMed Central CAS Google Scholar * Ito, M. et al. IMA peptides regulate
root nodulation and nitrogen homeostasis by providing iron according to internal nitrogen status. _Nat. Commun._ 15, 733 (2024). Article ADS PubMed PubMed Central CAS Google Scholar *
Bai, M. et al. Generation of a multiplex mutagenesis population via pooled CRISPR-Cas9 in soya bean. _Plant Biotechnol. J._ 18, 721–731 (2020). Article PubMed CAS Google Scholar * Song,
S. et al. Soybean seeds expressing feedback-insensitive cystathionine gamma-synthase exhibit a higher content of methionine. _J. Exp. Bot._ 64, 1917–1926 (2013). Article PubMed CAS Google
Scholar * Peng, W. T. et al. Magnesium promotes root nodulation through facilitation of carbohydrate allocation in soybean. _Physiol. Plant_ 163, 372–385 (2018). Article CAS Google
Scholar * Roschzttardtz, H., Conejero, G., Curie, C. & Mari, S. Identification of the endodermal vacuole as the iron storage compartment in the Arabidopsis embryo. _Plant Physiol_. 151,
1329–1338 (2009). Article PubMed PubMed Central CAS Google Scholar * Yoo, S. D., Cho, Y. H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient
gene expression analysis. _Nat. Protoc._ 2, 1565–1572 (2007). Article PubMed CAS Google Scholar * Wang, L. et al. A transcription factor of the NAC family regulates nitrate-induced
legume nodule senescence. _The New phytologist_ 238, 2113–2129 (2023). Article PubMed CAS Google Scholar * Song, W. Y. et al. A rice ABC transporter, OsABCC1, reduces arsenic
accumulation in the grain. _Proc. Natl. Acad. Sci. USA._ 111, 15699–15704 (2014). Article ADS PubMed PubMed Central CAS Google Scholar * Eide, D., Broderius, M., Fett, J. &
Guerinot, M. L. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. _Proc. Natl. Acad. Sci. USA._ 93, 5624–5628 (1996). Article ADS PubMed
PubMed Central CAS Google Scholar * Wang, L. L. et al. Single cell-type transcriptome profiling reveals genes that promote nitrogen fixation in the infected and uninfected cells of legume
nodules. _Plant Biotechnol. J._ 20, 616–618 (2022). Article PubMed PubMed Central CAS Google Scholar Download references ACKNOWLEDGEMENTS We thank Dr. Yong-Jia Zhong for providing
RFP-expressing rhizobia, Dr. Jian-Feng Ma for providing _fet3fet4_ mutant and stable isotope 57Fe, Dr. Xi Chen for providing _Δccc1_ mutant, Dr. Wen-Fei Wang for providing Nodulin 26
antibody. This work is financially supported by the National Natural Science Foundation of China (No. 32370284). AUTHOR INFORMATION Author notes * These authors contributed equally: Min
Zhou, Yuan Li, Xiao-Lei Yao. AUTHORS AND AFFILIATIONS * Haixia Institute of Science and Technology, Fujian Agriculture and Forestry University, Fuzhou, China Min Zhou, Yuan Li, Xiao-Lei Yao,
Jing Zhang, Sheng Liu, Hong-Rui Cao, Shuang Bai, Chun-Qu Chen, Dan-Xun Zhang, Ao Xu & Zhi-Chang Chen * State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and
Safety of Agro-products, Key Laboratory of Biotechnology in Plant Protection of MARA, Key Laboratory of Green Plant Protection of Zhejiang Province, Institute of Plant Virology, Ningbo
University, Ningbo, China Jia-Ning Lei & Qian-Zhuo Mao * State Key Laboratory of Agricultural Microbiology, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, China Yu
Zhou & De-Qiang Duanmu * Guangdong Provincial Key Laboratory of Plant Adaptation and Molecular Design, Innovative Center of Molecular Genetics and Evolution, School of Life Sciences,
Guangzhou University, Guangzhou, Guangdong, China Yue-Feng Guan Authors * Min Zhou View author publications You can also search for this author inPubMed Google Scholar * Yuan Li View author
publications You can also search for this author inPubMed Google Scholar * Xiao-Lei Yao View author publications You can also search for this author inPubMed Google Scholar * Jing Zhang View
author publications You can also search for this author inPubMed Google Scholar * Sheng Liu View author publications You can also search for this author inPubMed Google Scholar * Hong-Rui
Cao View author publications You can also search for this author inPubMed Google Scholar * Shuang Bai View author publications You can also search for this author inPubMed Google Scholar *
Chun-Qu Chen View author publications You can also search for this author inPubMed Google Scholar * Dan-Xun Zhang View author publications You can also search for this author inPubMed Google
Scholar * Ao Xu View author publications You can also search for this author inPubMed Google Scholar * Jia-Ning Lei View author publications You can also search for this author inPubMed
Google Scholar * Qian-Zhuo Mao View author publications You can also search for this author inPubMed Google Scholar * Yu Zhou View author publications You can also search for this author
inPubMed Google Scholar * De-Qiang Duanmu View author publications You can also search for this author inPubMed Google Scholar * Yue-Feng Guan View author publications You can also search
for this author inPubMed Google Scholar * Zhi-Chang Chen View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Z.C. conceived and designed the
experiments. M.Z. and Y.L. performed most of the experiments. X.Y. and Y.G. helped with the construction of the CRISPR-Cas9 materials. J.Z., S.L., and H.C. contributed to vector
constructions. S.B. contributed to symbiosome isolation. C.C. performed tobacco transient expression assay. D.Z. and A.X. performed yeast vacuolar isolation. J.L. and Q.M. performed the
immunoelectron microscopy. Y.Z. and D.D. helped with the separation of infected and uninfected cells. M.Z., Y.L., and Z.C. analyzed the data. Z.C. helped with microscopic observation and
wrote the manuscript. All authors discussed the results and commented on the manuscript. CORRESPONDING AUTHORS Correspondence to De-Qiang Duanmu, Yue-Feng Guan or Zhi-Chang Chen. ETHICS
DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. PEER REVIEW PEER REVIEW INFORMATION _Nature Communications_ thanks the anonymous, reviewer(s) for their
contribution to the peer review of this work. A peer review file is available. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in
published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION PEER REVIEW FILE DESCRIPTION OF ADDITIONAL SUPPLEMENTARY FILES SUPPLEMENTARY DATA 1
SUPPLEMENTARY DATA 2 SUPPLEMENTARY DATA 3 SUPPLEMENTARY DATA 4 REPORTING SUMMARY SOURCE DATA SOURCE DATA RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons
Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original
author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the
article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your
intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence,
visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Zhou, M., Li, Y., Yao, XL. _et al._ Inorganic nitrogen inhibits symbiotic
nitrogen fixation through blocking NRAMP2-mediated iron delivery in soybean nodules. _Nat Commun_ 15, 8946 (2024). https://doi.org/10.1038/s41467-024-53325-y Download citation * Received: 15
October 2023 * Accepted: 08 October 2024 * Published: 17 October 2024 * DOI: https://doi.org/10.1038/s41467-024-53325-y SHARE THIS ARTICLE Anyone you share the following link with will be
able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative