Play all audios:
ABSTRACT The correct sorting of nascent ribosomal proteins from the cytoplasm to the nucleus or to mitochondria for ribosome production poses a logistical challenge for cellular targeting
pathways. Here we report the discovery of a conserved mitochondrial avoidance segment (MAS) within the cytosolic ribosomal protein uS5 that resolves an evolutionary lethal conflict between
the nuclear and mitochondrial targeting machinery. MAS removal mistargets uS5 to the mitochondrial matrix and disrupts the assembly of the cytosolic ribosome. The resulting lethality can be
rescued by impairing mitochondrial import. We show that MAS triages nuclear targeting by disabling a cryptic mitochondrial targeting activity within uS5 and thereby prevents fatal capture by
mitochondria. Our findings identify MAS as an essential acquisition by the primordial eukaryote that reinforced organelle targeting fidelity while developing an endosymbiotic relationship
with its mitochondrial progenitor. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your
institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $29.99 / 30 days cancel any time Learn more Subscribe to this journal
Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices
may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support
SIMILAR CONTENT BEING VIEWED BY OTHERS COUPLING OF RIBOSOME BIOGENESIS AND TRANSLATION INITIATION IN HUMAN MITOCHONDRIA Article Open access 17 April 2025 DISTINCT MECHANISMS OF THE HUMAN
MITORIBOSOME RECYCLING AND ANTIBIOTIC RESISTANCE Article Open access 14 June 2021 A ROADMAP FOR RIBOSOME ASSEMBLY IN HUMAN MITOCHONDRIA Article Open access 11 July 2024 DATA AVAILABILITY All
data are presented in the main text and figures or supplementary information. Detailed protocols can be requested from the corresponding author. Proteomic data generated in this study have
been deposited in the ProteomeXchange Consortium via the PRIDE partner repository (https://www.proteomexchange.org) with the dataset identifier PXD035295 and are summarized in Supplementary
Table 3. The databases used in this study are the SGD (https://www.yeastgenome.org/) and the BLAST database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). All data supporting the findings of
this study are available from the corresponding author on reasonable request. Source data are provided with this paper. REFERENCES * Margulis, L. Symbiotic theory of the origin of eukaryotic
organelles; criteria for proof. _Symp. Soc. Exp. Biol._ 29, 21–38 (1975). Google Scholar * Youle, R. J. Mitochondria—striking a balance between host and endosymbiont. _Science_ 365,
eaaw9855 (2019). Article CAS PubMed Google Scholar * Muñoz-Gómez, S. A. et al. Site-and-branch-heterogeneous analyses of an expanded dataset favour mitochondria as sister to known
Alphaproteobacteria. _Nat. Ecol. Evol._ 6, 253–262 (2022). Article PubMed Google Scholar * Wiedemann, N. & Pfanner, N. Mitochondrial machineries for protein import and assembly.
_Annu. Rev. Biochem._ 86, 685–714 (2017). Article CAS PubMed Google Scholar * Bykov, Y. S. et al. Widespread use of unconventional targeting signals in mitochondrial ribosome proteins.
_EMBO J._ 41, e109519 (2022). Article CAS PubMed Google Scholar * Song, J., Herrmann, J. M. & Becker, T. Quality control of the mitochondrial proteome. _Nat. Rev. Mol. Cell Biol._
22, 54–70 (2021). Article CAS PubMed Google Scholar * von Heijne, G. Mitochondrial targeting sequences may form amphiphilic helices. _EMBO J._ 5, 1335–1342 (1986). Article Google
Scholar * Backes, S. et al. Tom70 enhances mitochondrial preprotein import efficiency by binding to internal targeting sequences. _J. Cell Biol._ 217, 1369–1382 (2018). Article CAS PubMed
PubMed Central Google Scholar * Stan, T. et al. Mitochondrial protein import: recognition of internal import signals of BCS1 by the TOM complex. _Mol. Cell. Biol._ 23, 2239–2250 (2003).
Article CAS PubMed PubMed Central Google Scholar * Rimmer, K. A. et al. Recognition of mitochondrial targeting sequences by the import receptors Tom20 and Tom22. _J. Mol. Biol._ 405,
804–818 (2011). Article CAS PubMed Google Scholar * Sirrenberg, C., Bauer, M. F., Guiard, B., Neupert, W. & Brunner, M. Import of carrier proteins into the mitochondrial inner
membrane mediated by Tim22. _Nature_ 384, 582–585 (1996). Article CAS PubMed Google Scholar * Steger, H. F. et al. Import of ADP/ATP carrier into mitochondria: two receptors act in
parallel. _J. Cell Biol._ 111, 2353–2363 (1990). Article CAS PubMed Google Scholar * Hines, V. et al. Protein import into yeast mitochondria is accelerated by the outer membrane protein
MAS70. _EMBO J._ 9, 3191–3200 (1990). Article CAS PubMed PubMed Central Google Scholar * Woellhaf, M. W., Hansen, K. G., Garth, C. & Herrmann, J. M. Import of ribosomal proteins
into yeast mitochondria1. _Biochem. Cell Biol._ 92, 489–498 (2014). Article CAS PubMed Google Scholar * Rout, M. P., Blobel, G. & Aitchison, J. D. A distinct nuclear import pathway
used by ribosomal proteins. _Cell_ 89, 715–725 (1997). Article CAS PubMed Google Scholar * Peña, C., Hurt, E. & Panse, V. G. Eukaryotic ribosome assembly, transport and quality
control. _Nat. Struct. Mol. Biol._ 24, 689–699 (2017). Article PubMed Google Scholar * Klinge, S. & Woolford, J. L. Ribosome assembly coming into focus. _Nat. Rev. Mol. Cell Biol._
20, 116–131 (2019). Article CAS PubMed PubMed Central Google Scholar * Pillet, B., Mitterer, V., Kressler, D. & Pertschy, B. Hold on to your friends: dedicated chaperones of
ribosomal proteins: dedicated chaperones mediate the safe transfer of ribosomal proteins to their site of pre-ribosome incorporation. _Bioessays_ 39, 1–12 (2017). Article CAS PubMed
Google Scholar * Pausch, P. et al. Co-translational capturing of nascent ribosomal proteins by their dedicated chaperones. _Nat. Commun._ 6, 7494 (2015). Article CAS PubMed Google
Scholar * Black, J. J., Musalgaonkar, S. & Johnson, A. W. Tsr4 is a cytoplasmic chaperone for the ribosomal protein Rps2 in _Saccharomyces cerevisiae_. _Mol. Cell. Biol._ 39, e00094-19
(2019). Article PubMed PubMed Central Google Scholar * Rössler, I. et al. Tsr4 and Nap1, two novel members of the ribosomal protein chaperOME. _Nucleic Acids Res._ 47, 6984–7002 (2019).
Article PubMed PubMed Central Google Scholar * Sardana, R. et al. The DEAH-box helicase Dhr1 dissociates U3 from the Pre-rRNA to promote formation of the central pseudoknot. _PLoS Biol._
13, 1002083 (2015). Article Google Scholar * Ben-Shem, A. et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. _Science_ 334, 1524–1529 (2011). Article CAS PubMed
Google Scholar * Coureux, P. D., Lazennec-Schurdevin, C., Bourcier, S., Mechulam, Y. & Schmitt, E. Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of
translation initiation. _Commun. Biol._ 3, 58 (2020). Article CAS PubMed PubMed Central Google Scholar * Nürenberg‐Goloub, E. et al. Molecular analysis of the ribosome recycling factor
ABCE 1 bound to the 30S post‐splitting complex. _EMBO J._ 39, e103788 (2020). Article PubMed PubMed Central Google Scholar * Jumper, J. et al. Highly accurate protein structure
prediction with AlphaFold. _Nature_ 596, 583–589 (2021). Article CAS PubMed PubMed Central Google Scholar * Evans, R. et al. Protein complex prediction with AlphaFold-Multimer. Preprint
at _bioRxiv_ https://doi.org/10.1101/2021.10.04.463034 (2022). * Türker, C. et al. B-fabric: the Swiss army knife for life sciences. In _Advances in Database Technology EDBT 2010 13th
International Conference on Extending Database Technology Proceedings_ 717–720 (Association for Computing Machinery, 2010). * Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic
and integrative analysis of large gene lists using DAVID bioinformatics resources. _Nat. Protoc._ 4, 44–57 (2009). Article CAS PubMed Google Scholar * Milkereit, P. et al. A Noc complex
specifically involved in the formation and nuclear export of ribosomal 40S subunits. _J. Biol. Chem._ 278, 4072–4081 (2003). Article CAS PubMed Google Scholar * Cabantous, S.,
Terwilliger, T. C. & Waldo, G. S. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. _Nat. Biotechnol._ 23, 102–107 (2005). Article
CAS PubMed Google Scholar * Ruan, L. et al. Cytosolic proteostasis through importing of misfolded proteins into mitochondria. _Nature_ 543, 443–446 (2017). Article CAS PubMed PubMed
Central Google Scholar * Kalderon, D., Roberts, B. L., Richardson, W. D. & Smith, A. E. A short amino acid sequence able to specify nuclear location. _Cell_ 39, 499–509 (1984). Article
CAS PubMed Google Scholar * van Leeuwen, J. et al. Systematic analysis of bypass suppression of essential genes. _Mol. Syst. Biol._ 16, e9828 (2020). Article CAS PubMed PubMed
Central Google Scholar * López-Llano, J., Campos, L. A. & Sancho, J. Alpha-helix stabilization by alanine relative to glycine: roles of polar and apolar solvent exposures and of
backbone entropy. _Proteins_ 64, 769–778 (2006). Article PubMed Google Scholar * Oborská-Oplová, M., Fischer, U., Altvater, M. & Panse, V. G. Eukaryotic ribosome assembly and
nucleocytoplasmic transport. _Methods Mol. Biol._ 2533, 99–126 (2022). Article PubMed PubMed Central Google Scholar * Moy, T. I. & Silver, P. A. Requirements for the nuclear export
of the small ribosomal subunit. _J. Cell Sci._ 115, 2985–2995 (2002). Article CAS PubMed Google Scholar * Fischer, U. et al. A non-canonical mechanism for Crm1-export cargo complex
assembly. _eLife_ 2015, 1–20 (2015). CAS Google Scholar * Altvater, M., Schütz, S., Chang, Y. & Panse, V. G. in _Methods in Cell Biology_ Vol. 122, 437–461 (Academic, 2014). *
Eisenberg-Bord, M. et al. Cnm1 mediates nucleus–mitochondria contact site formation in response to phospholipid levels. _J. Cell Biol._ 220, e202104100 (2021). Article CAS PubMed PubMed
Central Google Scholar * Weidberg, H. & Amon, A. MitoCPR-A surveillance pathway that protects mitochondria in response to protein import stress. _Science_ 360, eaan4146 (2018). Article
PubMed PubMed Central Google Scholar * Okreglak, V. & Walter, P. The conserved AAA-ATPase Msp1 confers organelle specificity to tail-anchored proteins. _Proc. Natl Acad. Sci. USA_
111, 8019–8024 (2014). Article CAS PubMed PubMed Central Google Scholar * Metzger, M. B., Scales, J. L., Dunklebarger, M. F., Loncarek, J. & Weissman, A. M. A protein quality
control pathway at the mitochondrial outer membrane. _eLife_ 9, e51065 (2020). Article CAS PubMed PubMed Central Google Scholar * Mårtensson, C. U. et al. Mitochondrial protein
translocation-associated degradation. _Nature_ 569, 679–683 (2019). Article PubMed Google Scholar * Wang, L. & Walter, P. Msp1/ATAD1 in protein quality control and regulation of
synaptic activities. _Annu. Rev. Cell Dev. Biol._ 36, 141–164 (2020). Article PubMed Google Scholar * Chen, Y. et al. Msp1/ATAD1 maintains mitochondrial function by facilitating the
degradation of mislocalized tail-anchored proteins. _EMBO J._ 33, 1548–1564 (2014). Article CAS PubMed PubMed Central Google Scholar * Plitzko, B. & Loesgen, S. Measurement of
oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in culture cells for assessment of the energy metabolism. _Bio-Protoc._ 8, e2850 (2018). Article CAS PubMed
PubMed Central Google Scholar * Song, J. & Becker, T. Fidelity of organellar protein targeting. _Curr. Opin. Cell Biol._ 75, 102071 (2022). Article CAS PubMed Google Scholar *
Juszkiewicz, S. & Hegde, R. S. Quality control of orphaned proteins. _Mol. Cell_ 71, 443–457 (2018). Article CAS PubMed PubMed Central Google Scholar * Hurt, E. C. & Schatz, G.
A cytosolic protein contains a cryptic mitochondrial targeting signal. _Nature_ 325, 499–503 (1987). Article CAS PubMed Google Scholar * Margulis, L. & Bermudes, D. Symbiosis as a
mechanism of evolution: status of cell symbiosis theory. _Symbiosis_ 1, 101–124 (1985). CAS PubMed Google Scholar * Lang, B. F., Gray, M. W. & Burger, G. Mitochondrial genome
evolution and the origin of eukaryotes. _Annu. Rev. Genet._ 33, 351–397 (1999). Article CAS PubMed Google Scholar * Sagan, L. On the origin of mitosing cells. _J. Theor. Biol._ 14,
255–274 (1967). Article CAS PubMed Google Scholar * Mills, D. B. et al. Eukaryogenesis and oxygen in Earth history. _Nat_. _Ecol. Evol._ 6, 520–532 (2022). Google Scholar * Bader, G. et
al. Assigning mitochondrial localization of dual localized proteins using a yeast bi-genomic mitochondrial-split-gfp. _eLife_ 9, 1–24 (2020). Article Google Scholar * Emanuelsson, O.
& von Heijne, G. Prediction of organellar targeting signals. _Biochim. Biophys. Acta_ 1541, 114–119 (2001). Article CAS PubMed Google Scholar * Zheng, N. & Gierasch, L. M. Signal
sequences: the same yet different. _Cell_ 86, 849–852 (1996). Article CAS PubMed Google Scholar * Zhang, X. & Shan, S. O. Fidelity of cotranslational protein targeting by the signal
recognition particle. _Annu. Rev. Biophys._ 43, 381–408 (2014). Article CAS PubMed PubMed Central Google Scholar * Hegde, R. S. & Zavodszky, E. Recognition and degradation of
mislocalized proteins in health and disease _Cold Spring Harb. Perspect. Biol._ 11, a033902 (2019). Article CAS PubMed PubMed Central Google Scholar * Longtine, M. S. et al. Additional
modules for versatile and economical PCR-based gene deletion and modification in _Saccharomyces cerevisiae_. _Yeast_ 14, 953–961 (1998). Article CAS PubMed Google Scholar * Cox, J. &
Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. _Nat. Biotechnol._ 26, 1367–1372 (2008).
Article CAS PubMed Google Scholar * Wolski, W., Grossmann, J. & Panse, C. protViz/SRMService. _GitHub_ https://github.com/protViz/SRMService (2018). * Meurer, M. et al. Genome-wide
C-SWAT library for high-throughput yeast genome tagging. _Nat. Methods_ 15, 598–600 (2018). Article CAS PubMed Google Scholar * Cohen, Y. & Schuldiner, M. Advanced methods for
high-throughput microscopy screening of genetically modified yeast libraries. _Methods Mol. Biol._ 781, 127–159 (2011). Article CAS PubMed Google Scholar * Tong, A. H. Y. & Boone, C.
Synthetic genetic array analysis in _Saccharomyces cerevisiae_. _Methods Mol. Biol._ 313, 171–192 (2006). CAS PubMed Google Scholar * Hanscho, M. et al. Nutritional requirements of the
BY series of _Saccharomyces cerevisiae_ strains for optimum growth. _FEMS Yeast Res._ 12, 796–808 (2012). Article CAS PubMed Google Scholar * Schindelin, J. et al. Fiji—an open source
platform for biological image analysis. _Nat. Methods_ 9, 676–682 (2012). Article CAS PubMed Google Scholar * Bareth, B. et al. Oms1 associates with cytochrome c oxidase assembly
intermediates to stabilize newly synthesized Cox1. _Mol. Biol. Cell_ 27, 1570–1580 (2016). Article CAS PubMed PubMed Central Google Scholar * Wittig, I., Braun, H. P. & Schägger, H.
Blue native PAGE. _Nat. Protoc._ 1, 418–428 (2006). Article CAS PubMed Google Scholar * Wenger, C., Oeljeklaus, S., Warscheid, B., Schneider, A. & Harsman, A. A trypanosomal
orthologue of an intermembrane space chaperone has a non-canonical function in biogenesis of the single mitochondrial inner membrane protein translocase. _PLoS Pathog._ 13, e1006550 (2017).
Article PubMed PubMed Central Google Scholar * Michel, E., Cucuzza, S., Mittl, P. R. E., Zerbe, O. & Plückthun, A. Improved repeat protein stability by combined consensus and
computational protein design. _Biochemistry_ 62, 318–329 (2023). Article CAS PubMed Google Scholar * Delano, W. L. PyMOL: an open-source molecular graphics tool. * Petterson, Eric. et
al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. _Protein Sci._ 30, 70–82 (2021). Article Google Scholar * Cherry, J. M. et al. Saccharomyces Genome
Database: the genomics resource of budding yeast. _Nucleic Acids Res._ 40, D700 (2012). Article CAS PubMed Google Scholar * Altschul, S. F., Gish, W., Miller, W., Myers, E. W. &
Lipman, D. J. Basic local alignment search tool. _J. Mol. Biol._ 215, 403–410 (1990). Article CAS PubMed Google Scholar * Weill, U. et al. Genome-wide SWAp-Tag yeast libraries for
proteome exploration. _Nat. Methods_ 15, 617–622 (2018). Article CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS We thank M. Peter, D. Rappaport and R. Li
for generously sharing yeast strains and plasmids. We thank R. Pillai, H. Hilbi, M. Seeger, H. Meyer, M. Pilhofer and all members of the Panse laboratory for enthusiastic discussions, the
Center for Microscopy and Image analysis, University of Zurich (ZMB, UZH) for maintaining the imaging equipment and the Functional Genomic Center Zurich (FGCZ) for proteomic analysis. We
thank C. Pena for performing a comparative analysis of ribosomal proteins, F. Willenborg for technical support during the maternity leave of M.O.-O. M.O.-O. was supported by a Boehringer
Ingelheim Fonds PhD fellowship, and a Pregnancy and Maternity Leave Compensation Grant from National Center of Competence in Research (NCCR) RNA and Disease. V.G.P. is supported by grants
from the Swiss National Science Foundation (SNF 188527), NCCR RNA and Disease (182880), Eidgenössische Technische Hochschule Zürich (ETHZ), Novartis Foundation, Olga Mayenfisch Stiftung and
a Starting Grant Award from the European Research Council (ERC) (EURIBIO260676). Work in the laboratory of A.S. was supported in part by NCCR RNA and Disease (205601) and by project grant
SNF 205200 both funded by the Swiss National Science Foundation. Research of P.R. was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s
Excellence Strategy EXC 2067/1-390729940, SFB1565 (P14), the ERC Advanced Grant MiXpress (ERCAdG no. 101095062) and the Max Planck Society. Work in the Schuldiner laboratory is supported by
the ERC CoG OnTarget (864068). M.S. is an Incumbent of the Dr. Gilbert Omenn and Martha Darling Professorial Chair in Molecular Genetics. Funding: the Swiss National Science Foundation
(VGP); NCCR in RNA and Disease (205601) (V.G.P. and A.S.); Novartis Science Foundation (V.G.P.); Olga Mayenfisch Stiftung (V.G.P.); ERC Starting Grant Award EURIBIO260676 (V.G.P.);
Boehringer Ingelheim Fonds PhD fellowship (M.O.-O.); Swiss National Science Foundation (SNF 205200) (A.S.); DFG, German Research Foundation, EXC 2067/1-390729940, SFB1565 (P.R.); ERC
Advanced Grant MiXpress (ERCAdG No. 101095062) (P.R.); ERC Consolidator Grant OnTarget (864068) (M.S.); and Max Planck Society (P.R.). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Institute
of Biochemistry, ETH Zurich, Zurich, Switzerland Michaela Oborská-Oplová * Institute of Medical Microbiology, University of Zurich, Zurich, Switzerland Michaela Oborská-Oplová, Alexander
Gregor Geiger, Purnima Klingauf-Nerurkar & Vikram Govind Panse * Department of Biochemistry, University of Zurich, Zurich, Switzerland Erich Michel * Department of Cellular Biochemistry,
University Medical Center Goettingen, Goettingen, Germany Sven Dennerlein & Peter Rehling * Department of Molecular Genetics, Weizmann Institute of Science, Rehovot, Israel Yury S.
Bykov & Maya Schuldiner * Department of Chemistry, Biochemistry and Pharmaceutical Sciences, University of Bern, Bern, Switzerland Simona Amodeo & André Schneider * Max-Planck
Institute for Multidisciplinary Sciences, Goettingen, Germany Peter Rehling * Cluster of Excellence ‘Multiscale Bioimaging: from Molecular Machines to Networks of Excitable Cells’,
University of Goettingen, Goettingen, Germany Peter Rehling * Faculty of Science, University of Zurich, Zurich, Switzerland Vikram Govind Panse Authors * Michaela Oborská-Oplová View author
publications You can also search for this author inPubMed Google Scholar * Alexander Gregor Geiger View author publications You can also search for this author inPubMed Google Scholar *
Erich Michel View author publications You can also search for this author inPubMed Google Scholar * Purnima Klingauf-Nerurkar View author publications You can also search for this author
inPubMed Google Scholar * Sven Dennerlein View author publications You can also search for this author inPubMed Google Scholar * Yury S. Bykov View author publications You can also search
for this author inPubMed Google Scholar * Simona Amodeo View author publications You can also search for this author inPubMed Google Scholar * André Schneider View author publications You
can also search for this author inPubMed Google Scholar * Maya Schuldiner View author publications You can also search for this author inPubMed Google Scholar * Peter Rehling View author
publications You can also search for this author inPubMed Google Scholar * Vikram Govind Panse View author publications You can also search for this author inPubMed Google Scholar
CONTRIBUTIONS Experimental design: M.O.-O., V.G.P., S.D., P.R., S.A., A.S. and M.S. Experiment execution: M.O.-O., P.K.-N., A.G.G., E.M., S.D., S.A. and Y.S.B. Data analysis: M.O.-O.,
A.G.G., E.M., S.D. and S.A. Supervision: V.G.P., A.S., P.R. and M.S. Writing—original draft: M.O.-O. and V.G.P. Writing—review and editing: M.O.-O., V.G.P., M.S., Y.S.B., P.R. and A.S.
CORRESPONDING AUTHOR Correspondence to Vikram Govind Panse. ETHICS DECLARATIONS COMPETING INTERESTS All authors declare that they have no competing interests. PEER REVIEW PEER REVIEW
INFORMATION _Nature Cell Biology_ thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available. ADDITIONAL INFORMATION
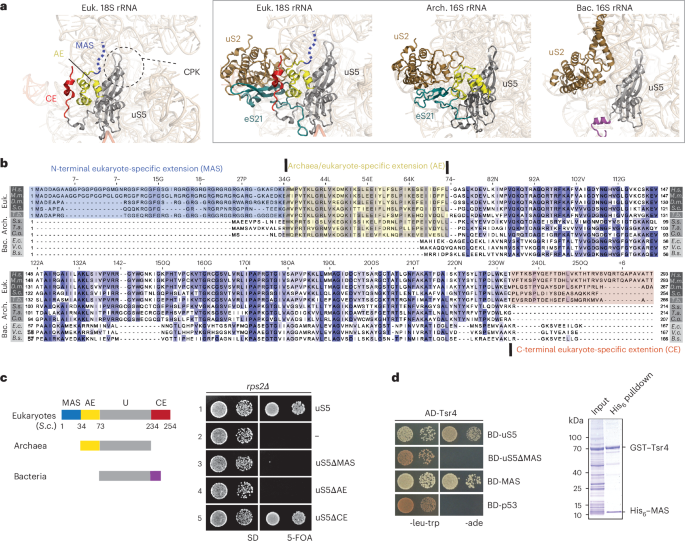
PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. EXTENDED DATA EXTENDED DATA FIG. 1 EUKARYOTE-SPECIFIC
SEGMENTS (ESSS) OF R-PROTEINS. Structures of 46 r-proteins from _S. cerevisiae_ (PDB ID: 4V7R) with marked ESSs based on the sequence alignment between _S. cerevisiae_ and archaeal species.
N-terminal ESSs in blue, insertions in green, and C-terminal ESSs in red. The shared sequence between _S. cerevisiae_ and archaea in yellow. EXTENDED DATA FIG. 2 MAS RECRUITS TSR4 FOR
HELIX-CHAPERONING. A, An AlphaFold generated multimer model of Tsr4-uS5 complex. The inter-residue distance errors in the model were low (pTM = 0.68), and the model’s confidence in the
positions of individual residues was high (pLDDT = 67.82). MAS (blue) and Tsr4 (pink) and the G128 residue (green) in the α-helix of the uS5 RNA-binding domain (grey) surrounded by Tsr4. B,
Western analysis of _tsr4∆_ cells expression uS5-GFP or uS5GA-GFP fusion proteins using indicated antibodies. Gsp1 was used as a loading control. Representative blot of _n_ = 3 independent
experiments is shown. C, Localisation of uS5-GFP or uS5GA-GFP fusions in _tsr4∆_ cells Scale bar = 5 µm. Representative images of _n_ = 3 biological replicates are shown. Source unprocessed
blots are available in source data. Source data EXTENDED DATA FIG. 3 MAS TRIAGES NUCLEAR TARGETING OF US5 FOR RIBOSOME ASSEMBLY ACROSS EUKARYOTES. A, Fluorescent images of split-GFP assay
performed with MTS-mCherry-GFP1-10 strain expressing _h_uS5 or _h_uS5ΔMAS fused to GFP11 fragment grown in selective 3% glycerol-containing media. Scale bar = 5 µm. B, Fluorescence imaging
of HeLa Flp-In T-Rex cells transiently expressing uS5-Venus or uS5∆MAS-Venus. Mitochondria were stained by Mitotracker Red. Scale bar = 20 μm. C, _upper panel_: Immunofluorescence assays of
_T. brucei_ cells expressing _Tb_uS5-HA or _Tb_uS5∆MAS-HA fusion proteins. ATOM40 was used as a mitochondrial marker. Scale bar = 5 µm. _lower panel_: Digitonin extractions of crude
mitochondrial fractions from _T. brucei_ cells expressing _Tb_uS5-HA or _Tb_uS5∆MAS-HA fusion proteins were analysed by immunoblots using the indicated antibodies. WC, whole cell extract; S,
cytosol-containing supernatant; P, mitochondria-enriched pellet. D, _Upper panel:_ Cartoon depicting uS5 domain organization from archaea _Thermoprotei archaeon. Lower panel:_ Fluorescence
imaging of wild-type yeast cells expressing uS5-GFP and AE-GFP from archaea _Thermoprotei archaeon_ (_Ta_). MTS-mCherry was used as a mitochondrial marker. The nucleus is indicated by a
white arrow. Scale bar = 5 µm. Source unprocessed blots are available in source data. Source data EXTENDED DATA FIG. 4 TRUNCATIONAL ANALYSIS OF MAS. Top: Cartoon depicting uS5GA domain
organization. Sequence of N-terminal MAS in blue, region shared between eukaryotes and archaea (AE) in yellow, universally conserved RNA-binding domain in grey, C-terminal
eukaryotic-specific extension (CE) in red. G128A point mutation in green. Bottom: _yrb2∆_ cells expressing C-terminal GFP fusions of uS5GA, uS5GA∆MAS, uS5GA∆N22, uS5GA∆N25 and uS5GA∆N28.
Representative images of _n_ = 3 biological replicates are shown. Mitochondria were stained by Mitotracker Red. Scale bar = 5 µm. EXTENDED DATA FIG. 5 FUNCTIONAL ANALYSIS OF US5ΔMAS CELLS.
A, _rps2∆ubx2∆_, _rps2∆msp1∆_, or _rps2∆cmn1∆_ shuffle strains transformed with empty vector or plasmids encoding uS5 or uS5∆MAS were spotted in 10-fold serial dilutions on selective SD and
5-FOA containing plates and grown at 30 °C for 3-8 days. B, Non-native extracts derived from mitochondria isolated from uS5 WT, uS5GA, and uS5GA∆MAS were separated by SDS-PAGE and subjected
to Western blotting analysis using indicated antibodies. C, mtDNA amplification of _COX1_ gene from _rps2∆_ or _rps2∆ tom70∆_ cells expressing indicated uS5 variants. Source unprocessed
blots are available in source data. Source data EXTENDED DATA FIG. 6 BI-GENOMIC SPLIT GFP ASSAY55. visualizes the mitochondrial fraction of any protein by using a yeast strain encoding
GFP1-10 within the mitochondrial (mt) DNA and 3xGFP11 fused to a protein of interest. A genomically tagged collection of all available cytosolic ribosomal proteins (RPs) was created using
the SWAp-Tag (SWAT) approach76. The collection was grown in YPD media, and representative images of _n_ = 3 individual experiments for r-proteins with mitochondrial fractions are shown along
with a no GFP11 control. mCherry fused to Su9MTS was used as a mitochondrial marker. Scale bar 5 µm. SUPPLEMENTARY INFORMATION REPORTING SUMMARY PEER REVIEW FILE SUPPLEMENTARY TABLES 1–4 MS
data and list of oligonucleotides. SOURCE DATA SOURCE DATA ALL Statistical source data. SOURCE DATA ALL Uncropped gels. RIGHTS AND PERMISSIONS Springer Nature or its licensor (e.g. a
society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript
version of this article is solely governed by the terms of such publishing agreement and applicable law. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Oborská-Oplová, M.,
Geiger, A.G., Michel, E. _et al._ An avoidance segment resolves a lethal nuclear–mitochondrial targeting conflict during ribosome assembly. _Nat Cell Biol_ 27, 336–346 (2025).
https://doi.org/10.1038/s41556-024-01588-4 Download citation * Received: 02 February 2024 * Accepted: 27 November 2024 * Published: 31 January 2025 * Issue Date: February 2025 * DOI:
https://doi.org/10.1038/s41556-024-01588-4 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative